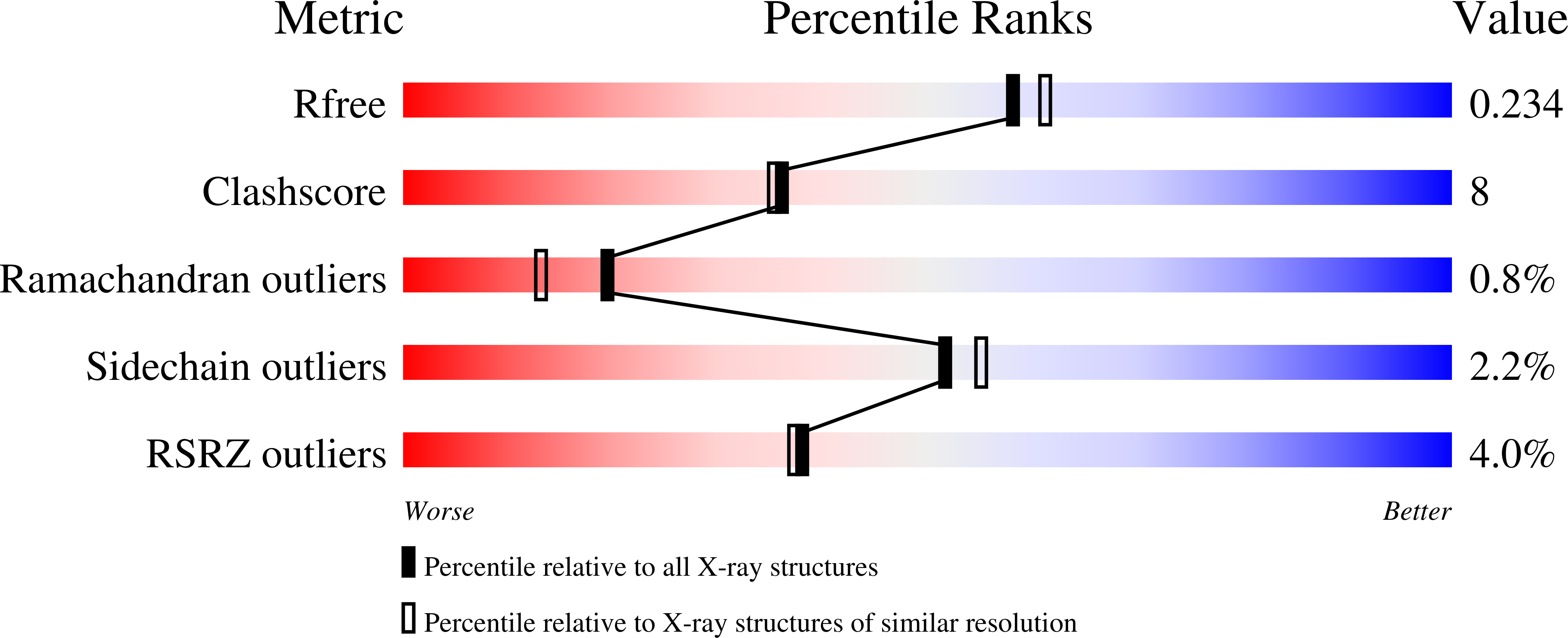
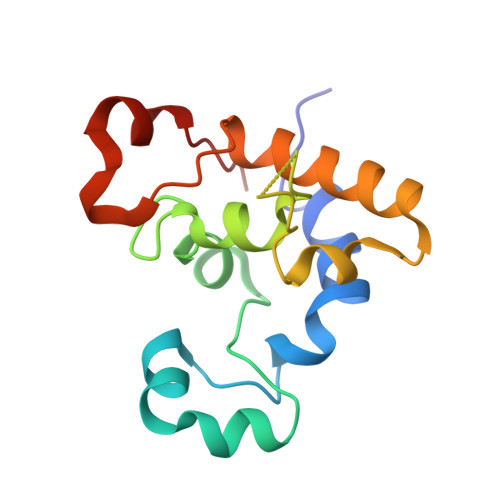
The structure of PccH from Geobacter sulfurreducens - a novel low reduction potential monoheme cytochrome essential for accepting electrons from an electrode.
Dantas, J.M., Campelo, L.M., Duke, N.E., Salgueiro, C.A., Pokkuluri, P.R.(2015) FEBS J 282: 2215-2231
- PubMed: 25786707
- DOI: https://doi.org/10.1111/febs.13269
- Primary Citation of Related Structures:
4RLR - PubMed Abstract:
The structure of cytochrome c (GSU3274) designated as PccH from Geobacter sulfurreducens was determined at a resolution of 2.0 Å. PccH is a small (15 kDa) cytochrome containing one c-type heme, found to be essential for the growth of G. sulfurreducens with respect to accepting electrons from graphite electrodes poised at -300 mV versus standard hydrogen electrode. with fumarate as the terminal electron acceptor. The structure of PccH is unique among the monoheme cytochromes described to date. The structural fold of PccH can be described as forming two lobes with the heme sandwiched in a cleft between the two lobes. In addition, PccH has a low reduction potential of -24 mV at pH 7, which is unusual for monoheme cytochromes. Based on difference in structure, together with sequence phylogenetic analysis, we propose that PccH can be regarded as a first characterized example of a new subclass of class I monoheme cytochromes. The low reduction potential of PccH may enable the protein to be redox active at the typically negative potential ranges encountered by G. sulfurreducens. Because PccH is predicted to be located in the periplasm of this bacterium, it could not be involved in the first step of accepting electrons from the electrode but is very likely involved in the downstream electron transport events in the periplasm.
Organizational Affiliation:
UCIBIO - REQUIMTE, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, Caparica, Portugal.