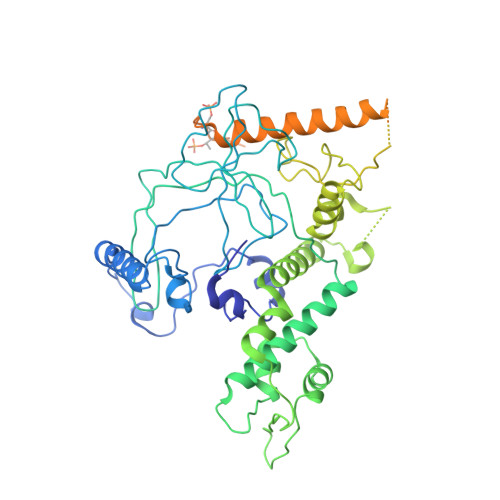
Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Wang, H., Curran, E.C., Hinds, T.R., Wang, E.H., Zheng, N.(2014) Cell Res 24: 1433-1444
- PubMed: 25412659
- DOI: https://doi.org/10.1038/cr.2014.148
- Primary Citation of Related Structures:
4RGW - PubMed Abstract:
The general transcription factor IID (TFIID) initiates RNA polymerase II-mediated eukaryotic transcription by nucleating pre-initiation complex formation at the core promoter of protein-encoding genes. TAF1, the largest integral subunit of TFIID, contains an evolutionarily conserved yet poorly characterized central core domain, whose specific mutation disrupts cell proliferation in the temperature-sensitive mutant hamster cell line ts13. Although the impaired TAF1 function in the ts13 mutant has been associated with defective transcriptional regulation of cell cycle genes, the mechanism by which TAF1 mediates transcription as part of TFIID remains unclear. Here, we present the crystal structure of the human TAF1 central core domain in complex with another conserved TFIID subunit, TAF7, which biochemically solubilizes TAF1. The TAF1-TAF7 complex displays an inter-digitated compact architecture, featuring an unexpected TAF1 winged helix (WH) domain mounted on top of a heterodimeric triple barrel. The single TAF1 residue altered in the ts13 mutant is buried at the junction of these two structural domains. We show that the TAF1 WH domain has intrinsic DNA-binding activity, which depends on characteristic residues that are commonly used by WH fold proteins for interacting with DNA. Importantly, mutations of these residues not only compromise DNA binding by TAF1, but also abrogate its ability to rescue the ts13 mutant phenotype. Together, our results resolve the structural organization of the TAF1-TAF7 module in TFIID and unveil a critical promoter-binding function of TAF1 in transcription regulation.
Organizational Affiliation:
1] Department of Pharmacology, Box 357280, University of Washington, Seattle, WA 98195, USA [2] Howard Hughes Medical Institute, Box 357280, University of Washington, Seattle, WA 98195, USA.