Structure of the N-terminal domain of the protein Expansion: an 'Expansion' to the Smad MH2
Beich-Frandsen, M., Aragon, E., Llimargas, M., Benach, J., Riera, A., Pous, J., Macias, M.J.(2015) Acta Crystallogr D Biol Crystallogr 71: 844-853
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2015) Acta Crystallogr D Biol Crystallogr 71: 844-853
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RE28239p | 242 | Drosophila melanogaster | Mutation(s): 0 Gene Names: CG13188 | 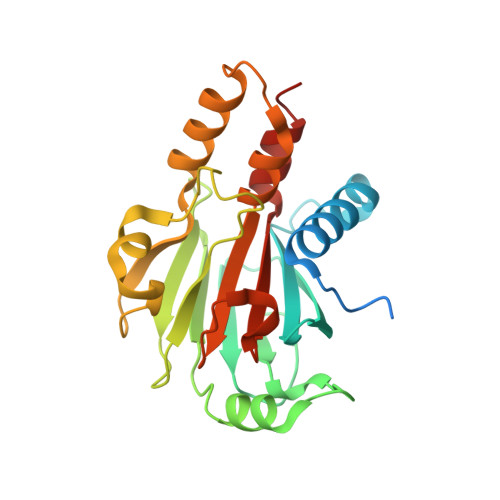 | |
UniProt | |||||
Find proteins for A1Z8R1 (Drosophila melanogaster) Explore A1Z8R1 Go to UniProtKB: A1Z8R1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A1Z8R1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 36.84 | α = 90 |
| b = 45.67 | β = 93.31 |
| c = 54.19 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| XDS | data reduction |