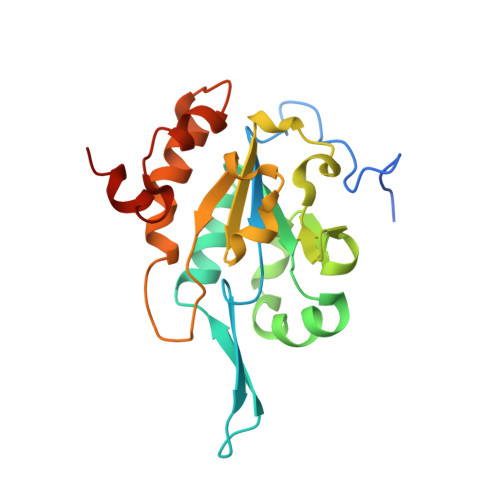
The structure of Tim50(164-361) suggests the mechanism by which Tim50 receives mitochondrial presequences.
Li, J., Sha, B.(2015) Acta Crystallogr F Struct Biol Commun 71: 1146-1151
- PubMed: 26323300
- DOI: https://doi.org/10.1107/S2053230X15013102
- Primary Citation of Related Structures:
4QQF - PubMed Abstract:
Mitochondrial preproteins are transported through the translocase of the outer membrane (TOM) complex. Tim50 and Tim23 then transfer preproteins with N-terminal targeting presequences through the intermembrane space (IMS) across the inner membrane. The crystal structure of the IMS domain of Tim50 [Tim50(164-361)] has previously been determined to 1.83 Å resolution. Here, the crystal structure of Tim50(164-361) at 2.67 Å resolution that was crystallized using a different condition is reported. Compared with the previously determined Tim50(164-361) structure, significant conformational changes occur within the protruding β-hairpin of Tim50 and the nearby helix A2. These findings indicate that the IMS domain of Tim50 exhibits significant structural plasticity within the putative presequence-binding groove, which may play important roles in the function of Tim50 as a receptor protein in the TIM complex that interacts with the presequence and multiple other proteins. More interestingly, the crystal packing indicates that helix A1 from the neighboring monomer docks into the putative presequence-binding groove of Tim50(164-361), which may mimic the scenario of Tim50 and the presequence complex. Tim50 may recognize and bind the presequence helix by utilizing the inner side of the protruding β-hairpin through hydrophobic interactions. Therefore, the protruding β-hairpin of Tim50 may play critical roles in receiving the presequence and recruiting Tim23 for subsequent protein translocations.
Organizational Affiliation:
Department of Cell, Developmental and Integrative Biology (CDIB), University of Alabama at Birmingham, Birmingham, AL 35294, USA.