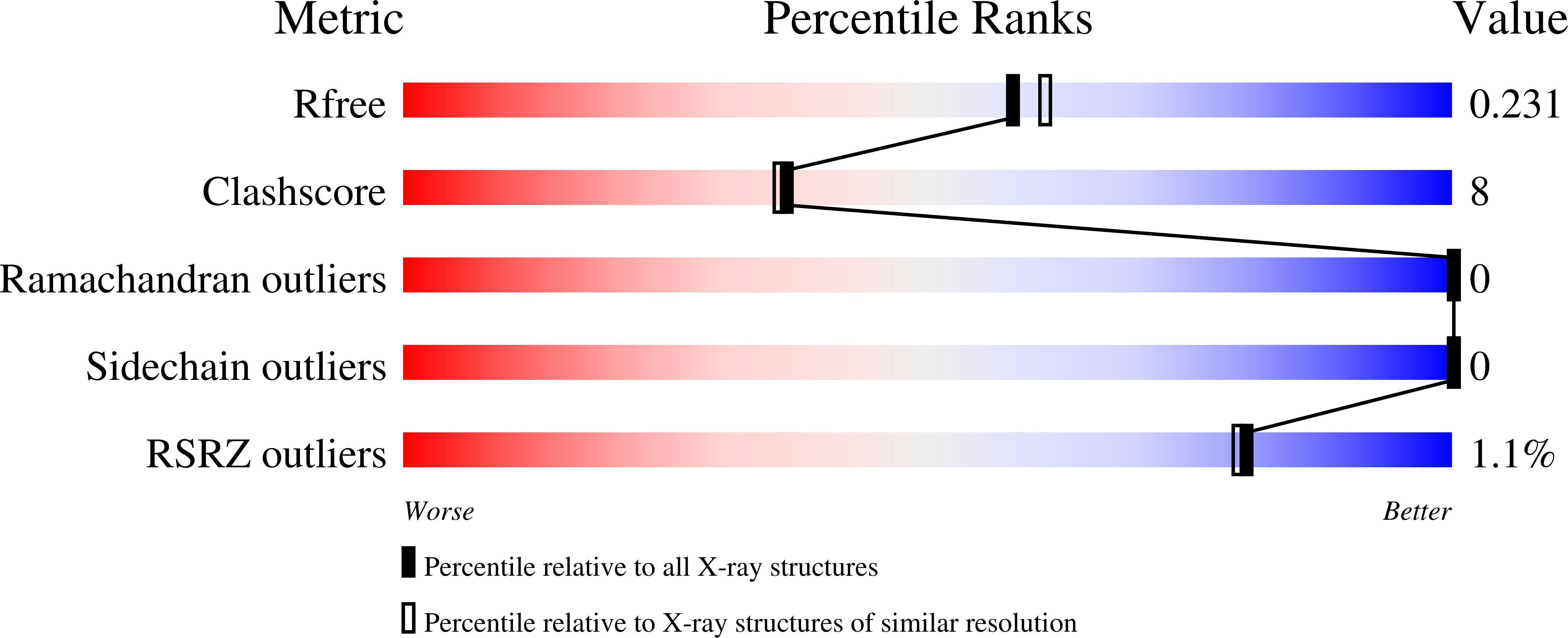
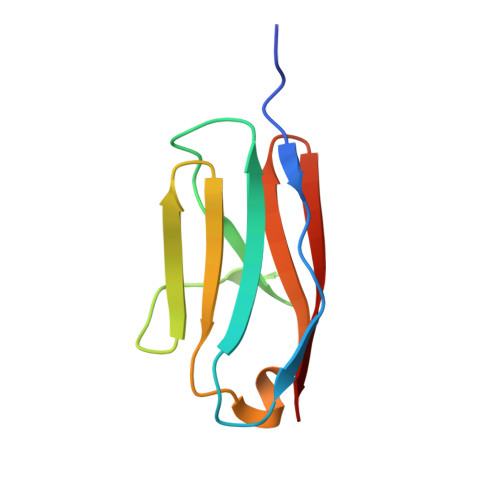
Exploration of the pathological potential of isolated mSNPs in titin: the cardiomyopathy-linked mutation T2580I
Bogomolovas, J., Labeit, S., Anderson, B., Williams, R., Lange, S., Simon, B., Khan, M.M., Rudolf, R., Bullard, B., Rigden, D.J., Granzier, H., Labiet, S., Mayans, O.(2016) Open Biol