Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Ortega, M.A., Velasquez, J.E., Garg, N., Zhang, Q., Joyce, R.E., Nair, S.K., van der Donk, W.A.(2014) ACS Chem Biol 9: 1718-1725
- PubMed: 24866416
- DOI: https://doi.org/10.1021/cb5002526
- Primary Citation of Related Structures:
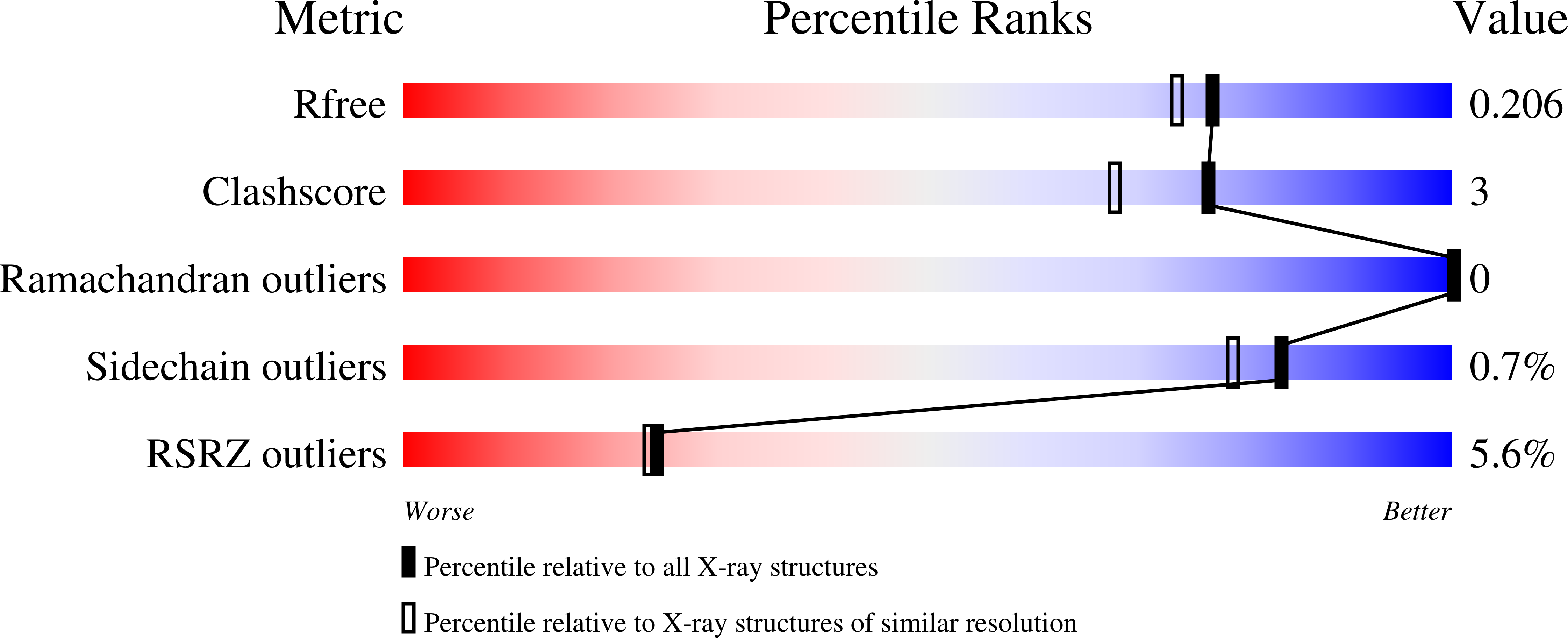
4QEC, 4QED - PubMed Abstract:
The final step in lanthipeptide biosynthesis involves the proteolytic removal of an N-terminal leader peptide. In the class I lanthipeptide epilancin 15X, this step is performed by the subtilisin-like serine peptidase ElxP. Bioinformatic, kinetic, and mass spectrometric analysis revealed that ElxP recognizes the stretch of amino acids DLNPQS located near the proteolytic cleavage site of its substrate, ElxA. When the ElxP recognition motif was inserted into the noncognate lanthipeptide precursor NisA, ElxP was able to proteolytically remove the leader peptide from NisA. Proteolytic removal of the leader peptide by ElxP during the biosynthesis of epilancin 15X exposes an N-terminal dehydroalanine on the core peptide of ElxA that hydrolyzes to a pyruvyl group. The short-chain dehydrogenase ElxO reduces the pyruvyl group to a lactyl moiety in the final step of epilancin 15X maturation. Using synthetic peptides, we also investigated the substrate specificity of ElxO and determined the 1.85 Å resolution X-ray crystal structure of the enzyme.
Organizational Affiliation:
Departments of Biochemistry and ‡Chemistry, and §the Howard Hughes Medical Institute, University of Illinois at Urbana-Champaign , 600 South Mathews Avenue, Urbana, Illinois 61801, United States.