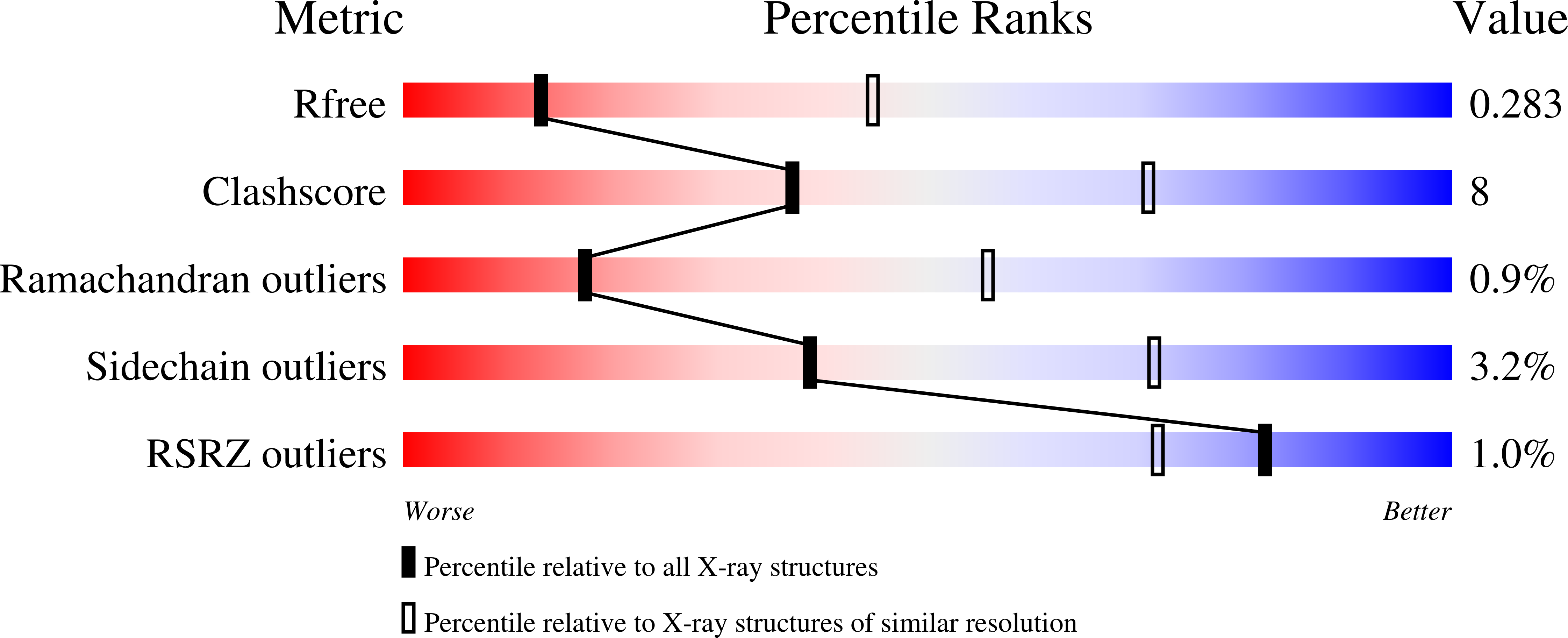
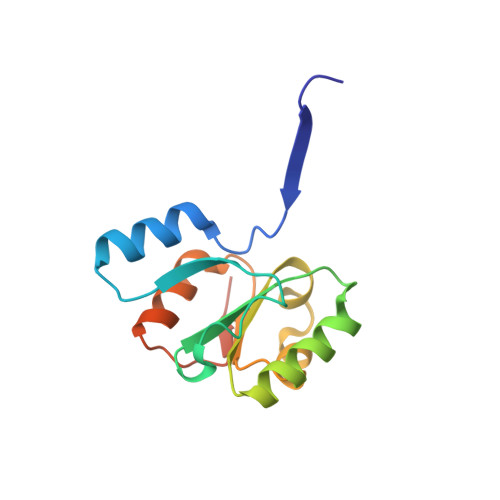
Evolutionary relationship of two ancient protein superfolds.
Farias-Rico, J.A., Schmidt, S., Hocker, B.(2014) Nat Chem Biol 10: 710-715
- PubMed: 25038785
- DOI: https://doi.org/10.1038/nchembio.1579
- Primary Citation of Related Structures:
4Q37 - PubMed Abstract:
Proteins are the molecular machines of the cell that fold into specific three-dimensional structures to fulfill their functions. To improve our understanding of how the structure and function of proteins arises, it is crucial to understand how evolution has generated the structural diversity we observe today. Classically, proteins that adopt different folds are considered to be nonhomologous. However, using state-of-the-art tools for homology detection, we found evidence of homology between proteins of two ancient and highly populated protein folds, the (βα)8-barrel and the flavodoxin-like fold. We detected a family of sequences that show intermediate features between both folds and determined what is to our knowledge the first representative crystal structure of one of its members, giving new insights into the evolutionary link of two of the earliest folds. Our findings contribute to an emergent vision where protein superfolds share common ancestry and encourage further approaches to complete the mapping of structure space onto sequence space.
Organizational Affiliation:
Max Planck Institute for Developmental Biology, Tübingen, Germany.