A conserved MCM single-stranded DNA binding element is essential for replication initiation.
Froelich, C.A., Kang, S., Epling, L.B., Bell, S.P., Enemark, E.J.(2014) Elife 3: e01993-e01993
- PubMed: 24692448
- DOI: https://doi.org/10.7554/eLife.01993
- Primary Citation of Related Structures:
4POF, 4POG - PubMed Abstract:
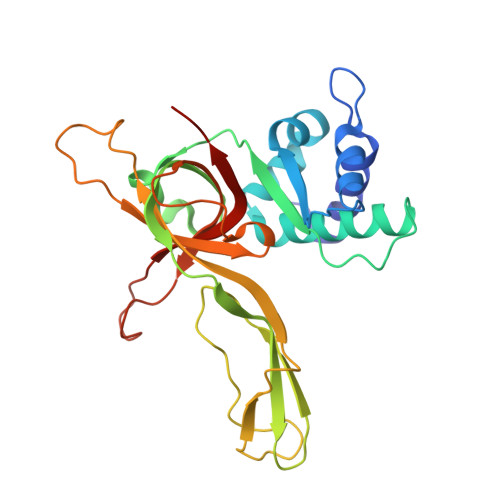
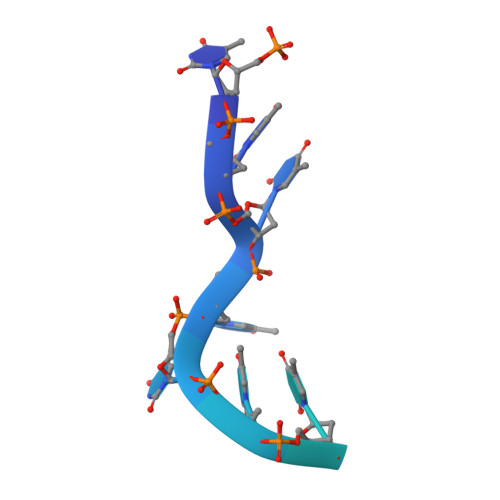
The ring-shaped MCM helicase is essential to all phases of DNA replication. The complex loads at replication origins as an inactive double-hexamer encircling duplex DNA. Helicase activation converts this species to two active single hexamers that encircle single-stranded DNA (ssDNA). The molecular details of MCM DNA interactions during these events are unknown. We determined the crystal structure of the Pyrococcus furiosus MCM N-terminal domain hexamer bound to ssDNA and define a conserved MCM-ssDNA binding motif (MSSB). Intriguingly, ssDNA binds the MCM ring interior perpendicular to the central channel with defined polarity. In eukaryotes, the MSSB is conserved in several Mcm2-7 subunits, and MSSB mutant combinations in S. cerevisiae Mcm2-7 are not viable. Mutant Mcm2-7 complexes assemble and are recruited to replication origins, but are defective in helicase loading and activation. Our findings identify an important MCM-ssDNA interaction and suggest it functions during helicase activation to select the strand for translocation. DOI: http://dx.doi.org/10.7554/eLife.01993.001.
Organizational Affiliation:
Department of Structural Biology, St Jude Children's Research Hospital, Memphis, United States.