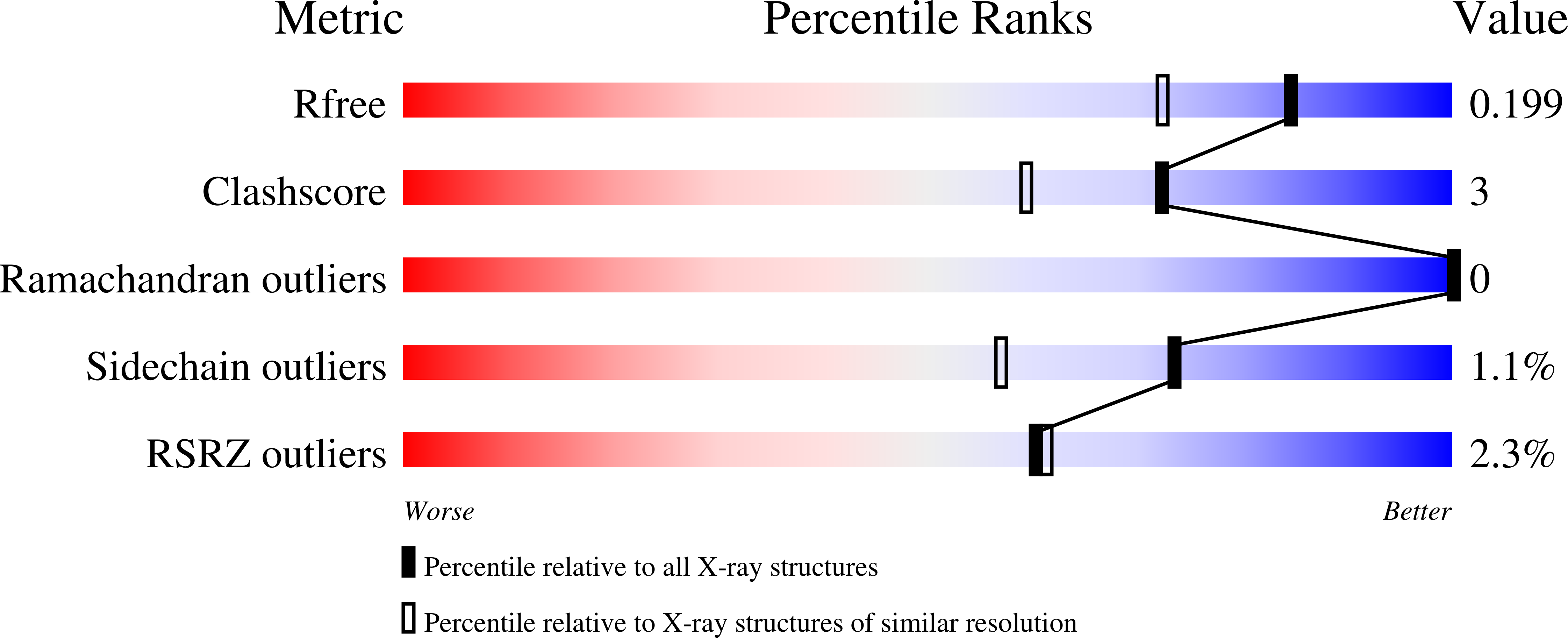
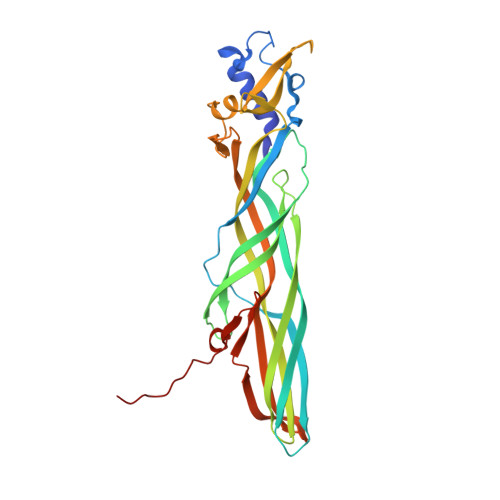
Crystal structure of Cry51Aa1: A potential novel insecticidal aerolysin-type beta-pore-forming toxin from Bacillus thuringiensis.
Xu, C., Chinte, U., Chen, L., Yao, Q., Meng, Y., Zhou, D., Bi, L.J., Rose, J., Adang, M.J., Wang, B.C., Yu, Z., Sun, M.(2015) Biochem Biophys Res Commun 462: 184-189
- PubMed: 25957471
- DOI: https://doi.org/10.1016/j.bbrc.2015.04.068
- Primary Citation of Related Structures:
4PKM - PubMed Abstract:
The structures of several Bacillus thuringiensis (Bt) insecticidal crystal proteins have been determined by crystallographic methods and a close relationship has been explicated between specific toxicities and conserved three-dimensional architectures. In this study, as a representative of the coleopteran- and hemipteran-specific Cry51A group, the complete structure of Cry51Aa1 protoxin has been determined by X-ray crystallography at 1.65 Å resolution. This is the first report of a coleopteran-active Bt insecticidal toxin with high structural similarity to the aerolysin-type β-pore forming toxins (β-PFTs). Moreover, study of featured residues and structural elements reveal their possible roles in receptor binding and pore formation events. This study provides new insights into the action of aerolysin-type β-PFTs from a structural perspective, and could be useful for the control of coleopteran and hemipteran insect pests in agricultures.
Organizational Affiliation:
State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei, 430070, PR China; Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, 30602, USA.