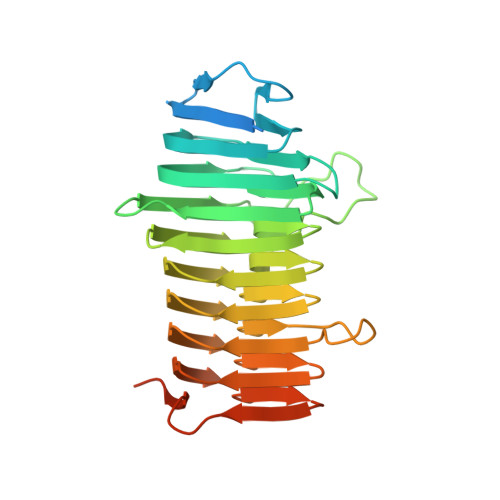
A new family of beta-helix proteins with similarities to the polysaccharide lyases.
Close, D.W., D'Angelo, S., Bradbury, A.R.(2014) Acta Crystallogr D Biol Crystallogr 70: 2583-2592
- PubMed: 25286843
- DOI: https://doi.org/10.1107/S1399004714015934
- Primary Citation of Related Structures:
4PEU, 4PHB - PubMed Abstract:
Microorganisms that degrade biomass produce diverse assortments of carbohydrate-active enzymes and binding modules. Despite tremendous advances in the genomic sequencing of these organisms, many genes do not have an ascribed function owing to low sequence identity to genes that have been annotated. Consequently, biochemical and structural characterization of genes with unknown function is required to complement the rapidly growing pool of genomic sequencing data. A protein with previously unknown function (Cthe_2159) was recently isolated in a genome-wide screen using phage display to identify cellulose-binding protein domains from the biomass-degrading bacterium Clostridium thermocellum. Here, the crystal structure of Cthe_2159 is presented and it is shown that it is a unique right-handed parallel β-helix protein. Despite very low sequence identity to known β-helix or carbohydrate-active proteins, Cthe_2159 displays structural features that are very similar to those of polysaccharide lyase (PL) families 1, 3, 6 and 9. Cthe_2159 is conserved across bacteria and some archaea and is a member of the domain of unknown function family DUF4353. This suggests that Cthe_2159 is the first representative of a previously unknown family of cellulose and/or acid-sugar binding β-helix proteins that share structural similarities with PLs. Importantly, these results demonstrate how functional annotation by biochemical and structural analysis remains a critical tool in the characterization of new gene products.
Organizational Affiliation:
Bioscience Division, Los Alamos National Laboratory, MS888, Los Alamos, NM 87545, USA.