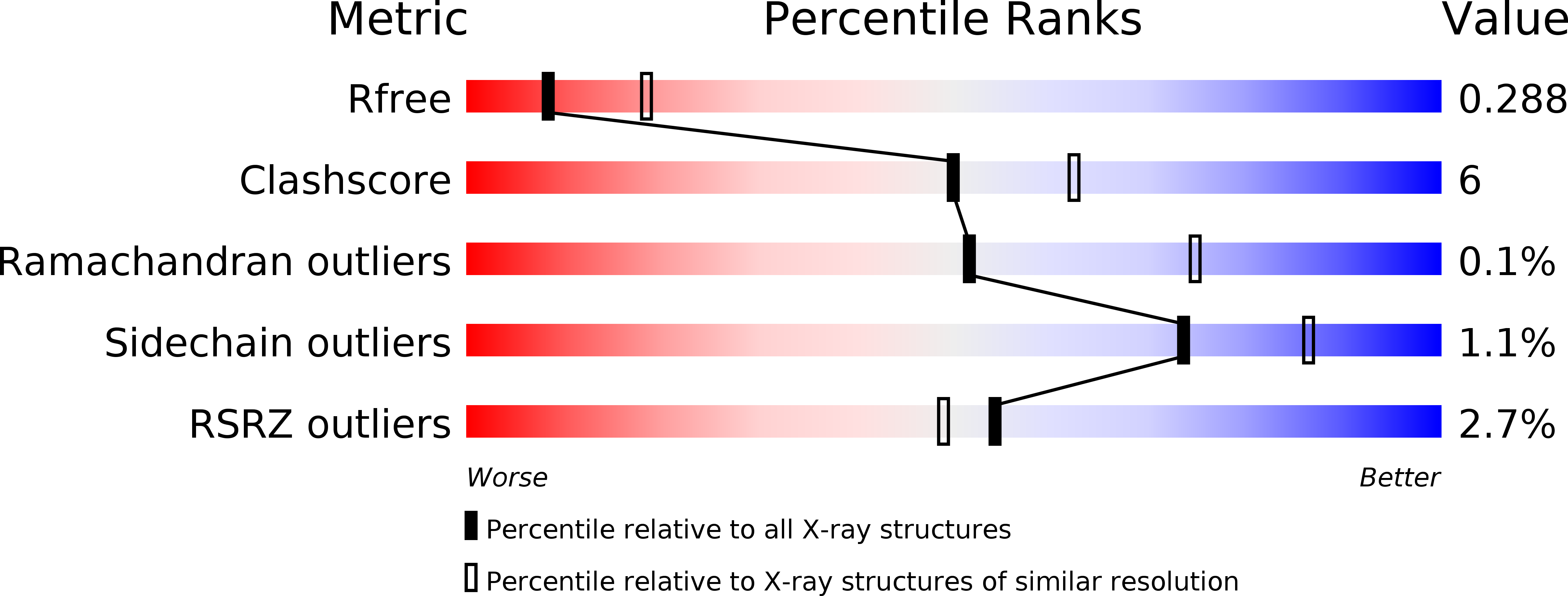
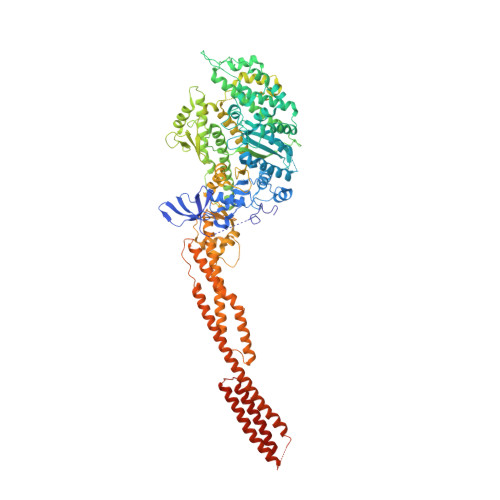
Crystal structure of the rigor-like human non-muscle myosin-2 motor domain.
Munnich, S., Pathan-Chhatbar, S., Manstein, D.J.(2014) FEBS Lett 588: 4754-4760
- PubMed: 25451231
- DOI: https://doi.org/10.1016/j.febslet.2014.11.007
- Primary Citation of Related Structures:
4PD3 - PubMed Abstract:
We determined the crystal structure of the motor domain of human non-muscle myosin 2B (NM-2B) in a nucleotide-free state and at a resolution of 2.8 Å. The structure shows the motor domain with an open active site and the large cleft that divides the 50 kDa domain in a closed state. Compared to other rigor-like myosin motor domain structures, our structure shows subtle but significant conformational changes in regions important for actin binding and mechanochemical coupling. Moreover, our crystal structure helps to rationalize the impact of myosin, heavy chain 9 (MYH9)-related disease mutations Arg709Cys and Arg709His on the kinetic and functional properties of NM-2B and of the closely related non-muscle myosin 2A (NM-2A).
Organizational Affiliation:
Institute for Biophysical Chemistry, Hannover Medical School, Hannover, Germany.