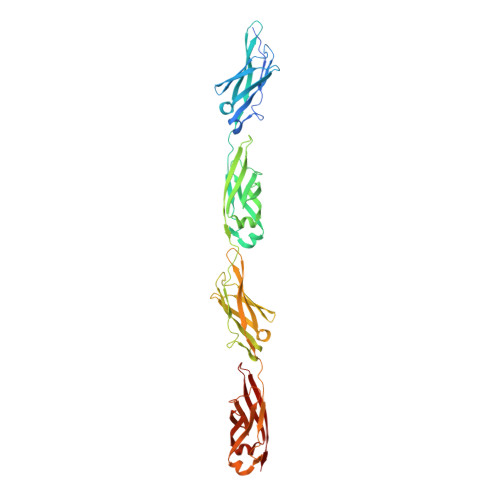Ca2+-stabilized adhesin helps an Antarctic bacterium reach out and bind ice.
Vance, T.D., Olijve, L.L., Campbell, R.L., Voets, I.K., Davies, P.L., Guo, S.(2014) Biosci Rep 34
- PubMed: 24892750
- DOI: https://doi.org/10.1042/BSR20140083
- Primary Citation of Related Structures:
4P99 - PubMed Abstract:
The large size of a 1.5-MDa ice-binding adhesin [MpAFP (Marinomonas primoryensis antifreeze protein)] from an Antarctic Gram-negative bacterium, M. primoryensis, is mainly due to its highly repetitive RII (Region II). MpAFP_RII contains roughly 120 tandem copies of an identical 104-residue repeat. We have previously determined that a single RII repeat folds as a Ca2+-dependent immunoglobulin-like domain. Here, we solved the crystal structure of RII tetra-tandemer (four tandem RII repeats) to a resolution of 1.8 Å. The RII tetra-tandemer reveals an extended (~190-Å × ~25-Å), rod-like structure with four RII-repeats aligned in series with each other. The inter-repeat regions of the RII tetra-tandemer are strengthened by Ca2+ bound to acidic residues. SAXS (small-angle X-ray scattering) profiles indicate the RII tetra-tandemer is significantly rigidified upon Ca2+ binding, and that the protein's solution structure is in excellent agreement with its crystal structure. We hypothesize that >600 Ca2+ help rigidify the chain of ~120 104-residue repeats to form a ~0.6 μm rod-like structure in order to project the ice-binding domain of MpAFP away from the bacterial cell surface. The proposed extender role of RII can help the strictly aerobic, motile bacterium bind ice in the upper reaches of the Antarctic lake where oxygen and nutrients are most abundant. Ca2+-induced rigidity of tandem Ig-like repeats in large adhesins might be a general mechanism used by bacteria to bind to their substrates and help colonize specific niches.
Organizational Affiliation:
*Protein Function Discovery Group and the Department of Biomedical and Molecular Sciences, Queen's University, Kingston, Ontario, Canada.