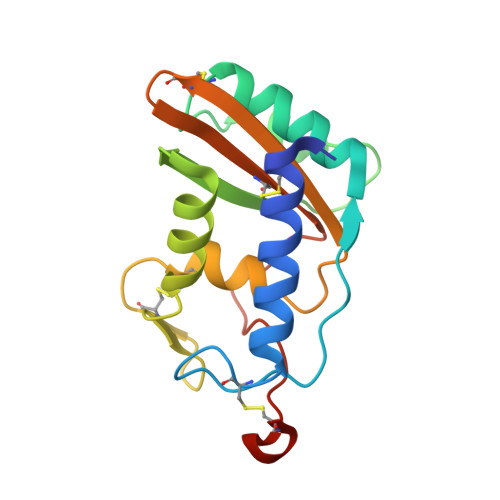
Schistosoma mansoni venom allergen-like protein 4 (SmVAL4) is a novel lipid-binding SCP/TAPS protein that lacks the prototypical CAP motifs.
Kelleher, A., Darwiche, R., Rezende, W.C., Farias, L.P., Leite, L.C., Schneiter, R., Asojo, O.A.(2014) Acta Crystallogr D Biol Crystallogr 70: 2186-2196
- PubMed: 25084337
- DOI: https://doi.org/10.1107/S1399004714013315
- Primary Citation of Related Structures:
4P27 - PubMed Abstract:
Schistosomiasis is a parasitic disease that affects over 200 million people. Vaccine candidates have been identified, including Schistosoma mansoni venom allergen-like proteins (SmVALs) from the SCP/TAPS (sperm-coating protein/Tpx/antigen 5/pathogenesis related-1/Sc7) superfamily. The first SmVAL structure, SmVAL4, was refined to a resolution limit of 2.16 Å. SmVAL4 has a unique structure that could not be predicted from homologous structures, with longer loops and an unusual C-terminal extension. SmVAL4 has the characteristic α/β-sandwich and central SCP/TAPS cavity. Furthermore, SmVAL4 has only one of the signature CAP cavity tetrad amino-acid residues and is missing the histidines that coordinate divalent cations such as Zn(2+) in other SCP/TAPS proteins. SmVAL4 has a cavity between α-helices 1 and 4 that was observed to bind lipids in tablysin-15, suggesting the ability to bind lipids. Subsequently, SmVAL4 was shown to bind cholesterol in vitro. Additionally, SmVAL4 was shown to complement the in vivo sterol-export phenotype of yeast mutants lacking their endogenous CAP proteins. Expression of SmVAL4 in yeast cells lacking endogenous CAP function restores the block in sterol export. These studies suggest an evolutionarily conserved lipid-binding function shared by CAP proteins such as SmVAL4 and yeast CAP proteins such as Pry1.
Organizational Affiliation:
National School of Tropical Medicine, Baylor College of Medicine, Houston, TX 77030, USA.