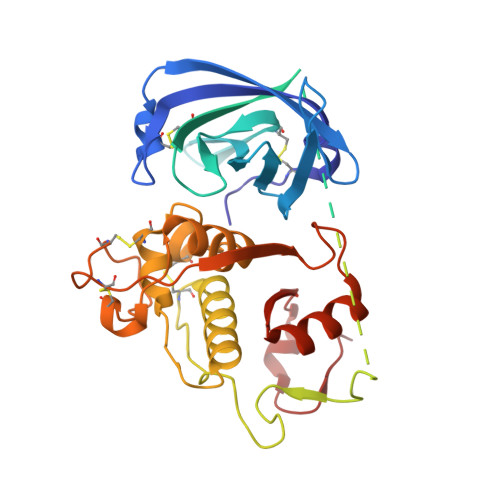
The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Rubach, J.K., Cui, G., Schneck, J.L., Taylor, A.N., Zhao, B., Smallwood, A., Nevins, N., Wisnoski, D., Thrall, S.H., Meek, T.D.(2012) Biochemistry 51: 7551-7568
- PubMed: 22928782
- DOI: https://doi.org/10.1021/bi300719b
- Primary Citation of Related Structures:
4OEL, 4OEM - PubMed Abstract:
We examined the cathepsin C-catalyzed hydrolysis of dipeptide substrates of the form Yaa-Xaa-AMC, using steady-state and pre-steady-state kinetic methods. The substrates group into three kinetic profiles based upon the broad range observed for k(cat)/K(a) and k(cat) values, pre-steady-state time courses, and solvent kinetic isotope effects (sKIEs). The dipeptide substrate Gly-Arg-AMC displayed large values for k(cat)/K(a) (1.6 ± 0.09 μM(-1) s(-1)) and k(cat) (255 ± 6 s(-1)), an inverse sKIE on k(cat)/K(a) ((D)(k(cat)/K(a)) = 0.6 ± 0.15), a modest, normal sKIE on k(cat) ((D)k(cat) = 1.6 ± 0.2), and immeasurable pre-steady-state kinetics, indicating an extremely fast pre-steady-state rate (>400 s(-1)). (Errors on fitted values are omitted in the text for clarity but may be found in Table 2.) These results conformed to a kinetic model where the acylation (k(ac)) and deacylation (k(dac)) half-reactions are very fast and similar in value. The second substrate type, Gly-Tyr-AMC and Ser-Tyr-AMC, the latter the subject of a comprehensive kinetic study (Schneck et al. (2008) Biochemistry 47, 8697-8710), were found to be less active substrates compared to Gly-Arg-AMC, with respective k(cat)/K(a) values of 0.49 ± 0.07 μM(-1 )s(-1) and 5.3 ± 0.5 μM(-1 )s(-1), and k(cat) values of 28 ± 1 s(-1) and 25 ± 0.5 s(-1). Solvent kinetic isotope effects for Ser-Tyr-AMC were found to be inverse for k(cat)/K(a) ((D)(k(cat)/K(a)) = 0.74 ± 0.05) and normal for k(cat) ((D)k(cat) = 2.3 ± 0.1) but unlike Gly-Arg-AMC, pre-steady-state kinetics of Gly-Tyr-AMC and Ser-Tyr-AMC were measurable and characterized by a single-exponential burst, with fast transient rates (490 s(-1) and 390 s(-1), respectively), from which it was determined that k(ac) ≫ k(dac) ∼ k(cat). The third substrate type, Gly-Ile-AMC, gave very low values of k(cat)/K(a) (0.0015 ± 0.0001 μM(-1) s(-1)) and k(cat) (0.33 ± 0.02 s(-1)), no sKIEs, ((D)(k(cat)/K(a)) = 1.05 ± 0.5 and (D)k(cat) = 1.06 ± 0.4), and pre-steady-state kinetics exhibited a discernible, but negligible, transient phase. For this third class of substrate, kinetic modeling was consistent with a mechanism in which k(dac) > k(ac) ∼ k(cat), and for which an isotope-insensitive step in the acylation half-reaction is the slowest. The combined results of these studies suggested that the identity of the amino acid at the P(1) position of the substrate is the main determinant of catalysis. On the basis of these kinetic data, together with crystallographic studies of substrate analogues and molecular dynamics analysis with models of acyl-enzyme intermediates, we present a catalytic model derived from the relative rates of the acylation vs deacylation half-reactions of cathepsin C. The chemical steps of catalysis are proposed to be dependent upon the conformational freedom of the amino acid substituents for optimal alignment for thiolation (acylation) or hydrolysis (deacylation). These studies suggest ideas for inhibitor design for papain-family cysteine proteases and strategies to progress drug discovery for other classes of disease-relevant cysteine proteases.
Organizational Affiliation:
Department of Biological Reagents and Assay Development, GlaxoSmithKline Pharmaceuticals, 1250 South Collegeville Road, Collegeville, PA 19426, USA.