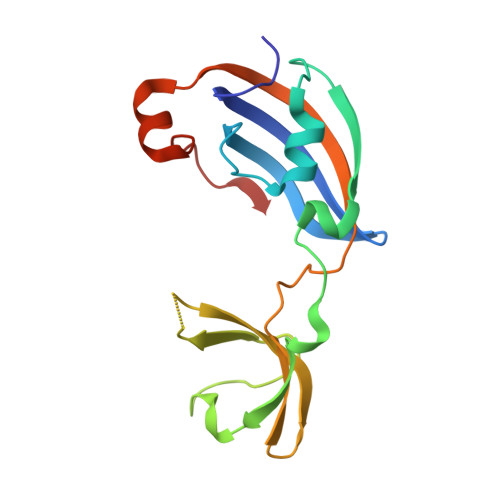
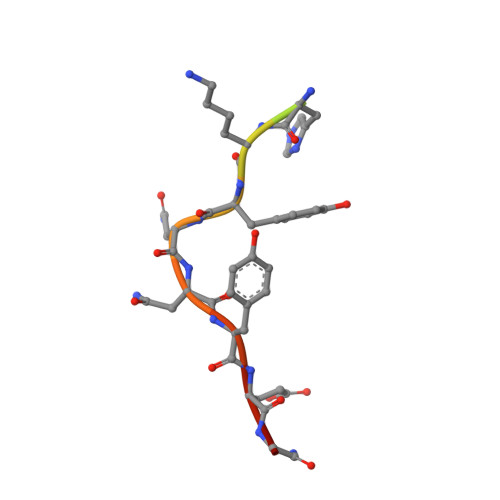
Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
Quistgaard, E.M., Weininger, U., Ural-Blimke, Y., Modig, K., Nordlund, P., Akke, M., Low, C.(2016) BMC Biol 14: 82-82
- PubMed: 27664121
- DOI: https://doi.org/10.1186/s12915-016-0300-3
- Primary Citation of Related Structures:
4ODK, 4ODL, 4ODM, 4ODN, 4ODO, 4ODP, 4ODQ, 4ODR - PubMed Abstract:
Peptidyl-prolyl isomerases (PPIases) catalyze cis/trans isomerization of peptidyl-prolyl bonds, which is often rate-limiting for protein folding. SlyD is a two-domain enzyme containing both a PPIase FK506-binding protein (FKBP) domain and an insert-in-flap (IF) chaperone domain. To date, the interactions of these domains with unfolded proteins have remained rather obscure, with structural information on binding to the FKBP domain being limited to complexes involving various inhibitor compounds or a chemically modified tetrapeptide.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Scheeles väg 2, SE-17177, Stockholm, Sweden.