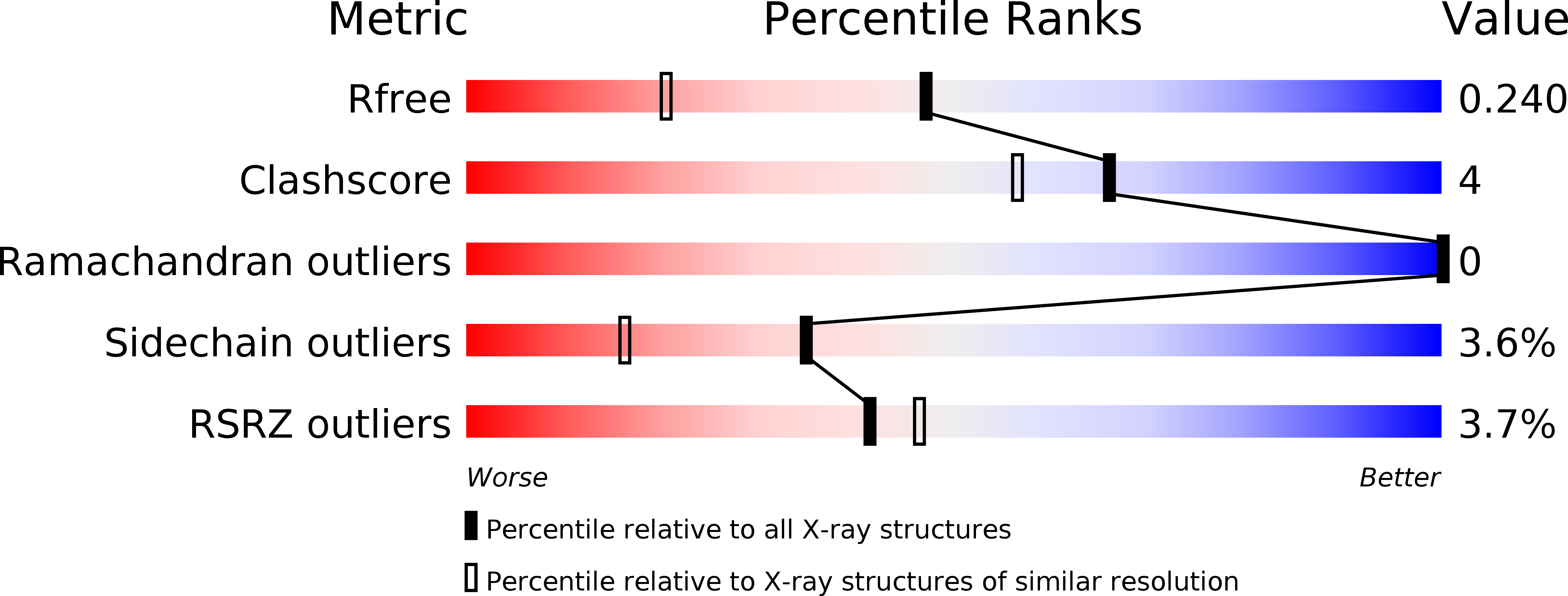
The condensin component NCAPG2 regulates microtubule-kinetochore attachment through recruitment of Polo-like kinase 1 to kinetochores.
Kim, J.H., Shim, J., Ji, M.J., Jung, Y., Bong, S.M., Jang, Y.J., Yoon, E.K., Lee, S.J., Kim, K.G., Kim, Y.H., Lee, C., Lee, B.I., Kim, K.T.(2014) Nat Commun 5: 4588-4588
- PubMed: 25109385
- DOI: https://doi.org/10.1038/ncomms5588
- Primary Citation of Related Structures:
4O9W - PubMed Abstract:
The early event of microtubule-kinetochore attachment is a critical stage for precise chromosome segregation. Here we report that NCAPG2, which is a component of the condensin II complex, mediates chromosome segregation through microtubule-kinetochore attachment by recruiting PLK1 to prometaphase kinetochores. NCAPG2 colocalizes with PLK1 at prometaphase kinetochores and directly interacts with the polo-box domain (PBD) of PLK1 via its highly conserved C-terminal region. In both humans and Caenorhabditis elegans, when NCAPG2 is depleted, the attachment of the spindle to the kinetochore is loosened and misoriented. This is caused by the disruption of PLK1 localization to the kinetochore and by the decreased phosphorylation of its kinetochore substrate, BubR1. In addition, the crystal structure of the PBD of PLK1, in complex with the C-terminal region of NCAPG2, (1007)VLS-pT-L(1011), exhibits structural conservation of PBD-phosphopeptides, suggesting that the regulation of NCAPG2 function is phosphorylation-dependent. These findings suggest that NCAPG2 plays an important role in regulating proper chromosome segregation through a functional interaction with PLK1 during mitosis.
Organizational Affiliation:
1] Research Institute, National Cancer Center, Goyang, Gyeonggi 410-769, Republic of Korea [2].