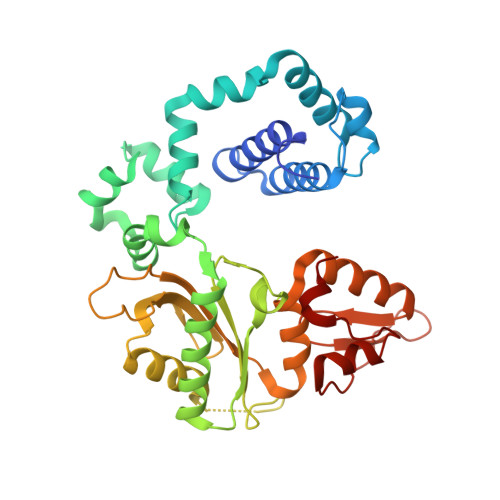
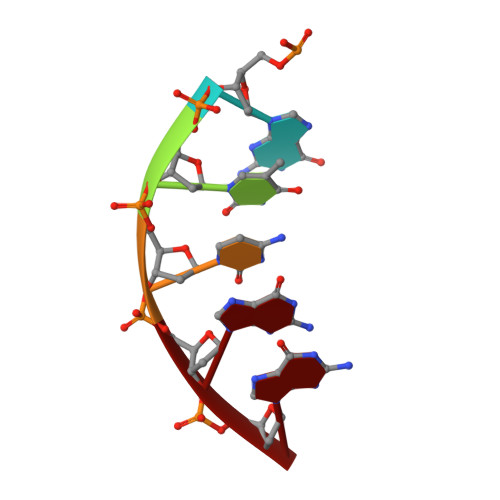
Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair.
Caglayan, M., Batra, V.K., Sassa, A., Prasad, R., Wilson, S.H.(2014) Nat Struct Mol Biol 21: 497-499
- PubMed: 24777061
- DOI: https://doi.org/10.1038/nsmb.2818
- Primary Citation of Related Structures:
4O9M - PubMed Abstract:
DNA polymerase β (pol β) lyase removal of 5'-deoxyribose phosphate (5'-dRP) from base excision repair (BER) intermediates is critical in mammalian BER involving the abasic site. We found that pol β also removes 5'-adenylated dRP from BER intermediates after abortive ligation. The crystal structure of a human pol β-DNA complex showed the 5'-AMP-dRP group positioned in the lyase active site. Pol β expression rescued methyl methanesulfonate sensitivity in aprataxin (hnt3)- and FEN1 (rad27)-deficient yeast.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, North Carolina, USA.