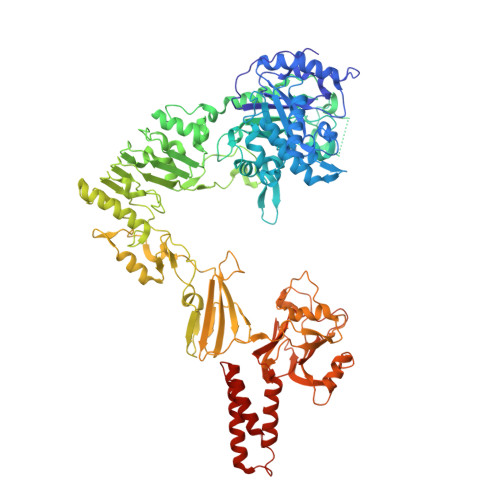
Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Trastoy, B., Lomino, J.V., Pierce, B.G., Carter, L.G., Gunther, S., Giddens, J.P., Snyder, G.A., Weiss, T.M., Weng, Z., Wang, L.X., Sundberg, E.J.(2014) Proc Natl Acad Sci U S A 111: 6714-6719
- PubMed: 24753590
- DOI: https://doi.org/10.1073/pnas.1322908111
- Primary Citation of Related Structures:
4NUY, 4NUZ - PubMed Abstract:
To evade host immune mechanisms, many bacteria secrete immunomodulatory enzymes. Streptococcus pyogenes, one of the most common human pathogens, secretes a large endoglycosidase, EndoS, which removes carbohydrates in a highly specific manner from IgG antibodies. This modification renders antibodies incapable of eliciting host effector functions through either complement or Fc γ receptors, providing the bacteria with a survival advantage. On account of this antibody-specific modifying activity, EndoS is being developed as a promising injectable therapeutic for autoimmune diseases that rely on autoantibodies. Additionally, EndoS is a key enzyme used in the chemoenzymatic synthesis of homogenously glycosylated antibodies with tailored Fc γ receptor-mediated effector functions. Despite the tremendous utility of this enzyme, the molecular basis of EndoS specificity for, and processing of, IgG antibodies has remained poorly understood. Here, we report the X-ray crystal structure of EndoS and provide a model of its encounter complex with its substrate, the IgG1 Fc domain. We show that EndoS is composed of five distinct protein domains, including glycosidase, leucine-rich repeat, hybrid Ig, carbohydrate binding module, and three-helix bundle domains, arranged in a distinctive V-shaped conformation. Our data suggest that the substrate enters the concave interior of the enzyme structure, is held in place by the carbohydrate binding module, and that concerted conformational changes in both enzyme and substrate are required for subsequent antibody deglycosylation. The EndoS structure presented here provides a framework from which novel endoglycosidases could be engineered for additional clinical and biotechnological applications.
Organizational Affiliation:
Institute of Human Virology and Departments of Biochemistry and Molecular Biology, Medicine, and Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, MD 21201.