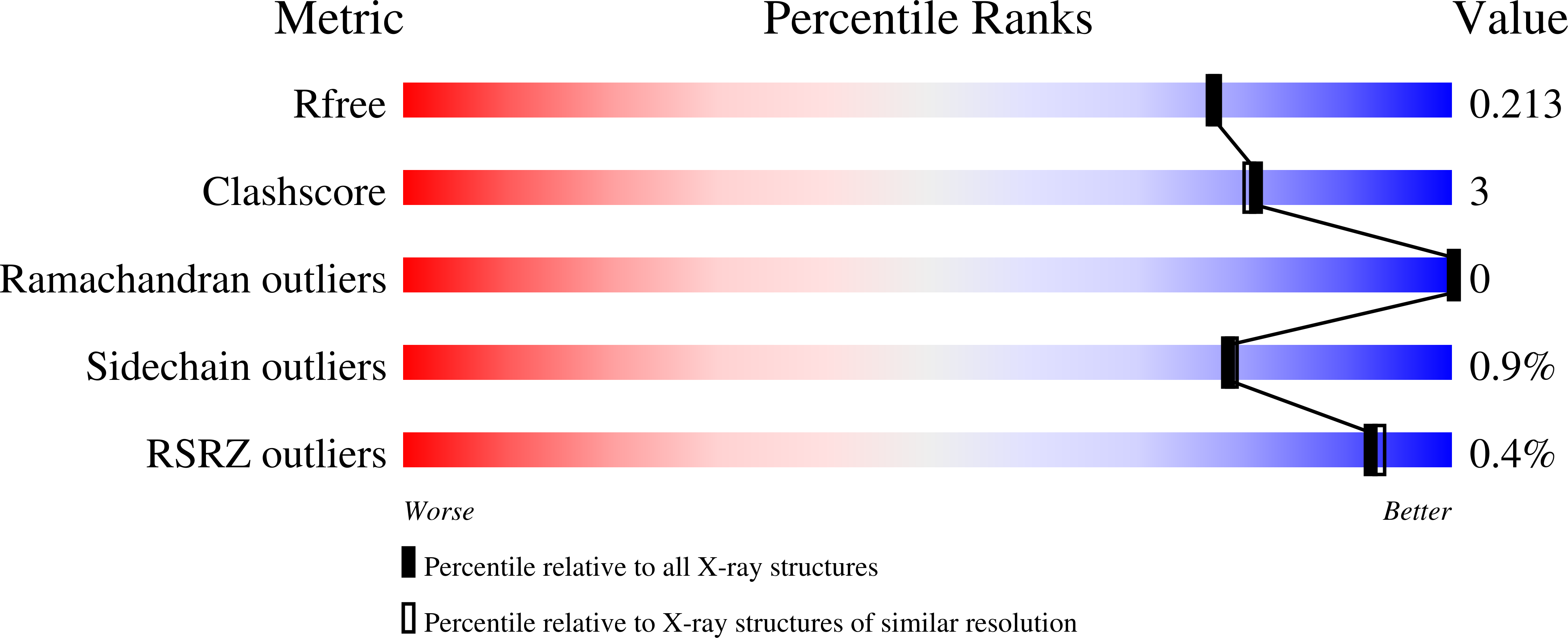
Evolutionary Diversification of Host-Targeted Bartonella Effectors Proteins Derived from a Conserved FicTA Toxin-Antitoxin Module.
Schirmer, T., de Beer, T.A.P., Tamegger, S., Harms, A., Dietz, N., Dranow, D.M., Edwards, T.E., Myler, P.J., Phan, I., Dehio, C.(2021) Microorganisms 9
- PubMed: 34442725
- DOI: https://doi.org/10.3390/microorganisms9081645
- Primary Citation of Related Structures:
4LU4, 4M16, 4N67, 4NPS, 4PY3, 4WGJ, 4XI8 - PubMed Abstract:
Proteins containing a FIC domain catalyze AMPylation and other post-translational modifications (PTMs). In bacteria, they are typically part of FicTA toxin-antitoxin modules that control conserved biochemical processes such as topoisomerase activity, but they have also repeatedly diversified into host-targeted virulence factors. Among these, Bartonella effector proteins (Beps) comprise a particularly diverse ensemble of FIC domains that subvert various host cellular functions. However, no comprehensive comparative analysis has been performed to infer molecular mechanisms underlying the biochemical and functional diversification of FIC domains in the vast Bep family. Here, we used X-ray crystallography, structural modelling, and phylogenetic analyses to unravel the expansion and diversification of Bep repertoires that evolved in parallel in three Bartonella lineages from a single ancestral FicTA toxin-antitoxin module. Our analysis is based on 99 non-redundant Bep sequences and nine crystal structures. Inferred from the conservation of the FIC signature motif that comprises the catalytic histidine and residues involved in substrate binding, about half of them represent AMP transferases. A quarter of Beps show a glutamate in a strategic position in the putative substrate binding pocket that would interfere with triphosphate-nucleotide binding but may allow binding of an AMPylated target for deAMPylation or another substrate to catalyze a distinct PTM. The β-hairpin flap that registers the modifiable target segment to the active site exhibits remarkable structural variability. The corresponding sequences form few well-defined groups that may recognize distinct target proteins. The binding of Beps to promiscuous FicA antitoxins is well conserved, indicating a role of the antitoxin to inhibit enzymatic activity or to serve as a chaperone for the FIC domain before translocation of the Bep into host cells. Taken together, our analysis indicates a remarkable functional plasticity of Beps that is mostly brought about by structural changes in the substrate pocket and the target dock. These findings may guide future structure-function analyses of the highly versatile FIC domains.
Organizational Affiliation:
Biozentrum, University of Basel, 4056 Basel, Switzerland.