Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor.
Lu, L., Liu, Q., Zhu, Y., Chan, K.H., Qin, L., Li, Y., Wang, Q., Chan, J.F., Du, L., Yu, F., Ma, C., Ye, S., Yuen, K.Y., Zhang, R., Jiang, S.(2014) Nat Commun 5: 3067-3067
- PubMed: 24473083
- DOI: https://doi.org/10.1038/ncomms4067
- Primary Citation of Related Structures:
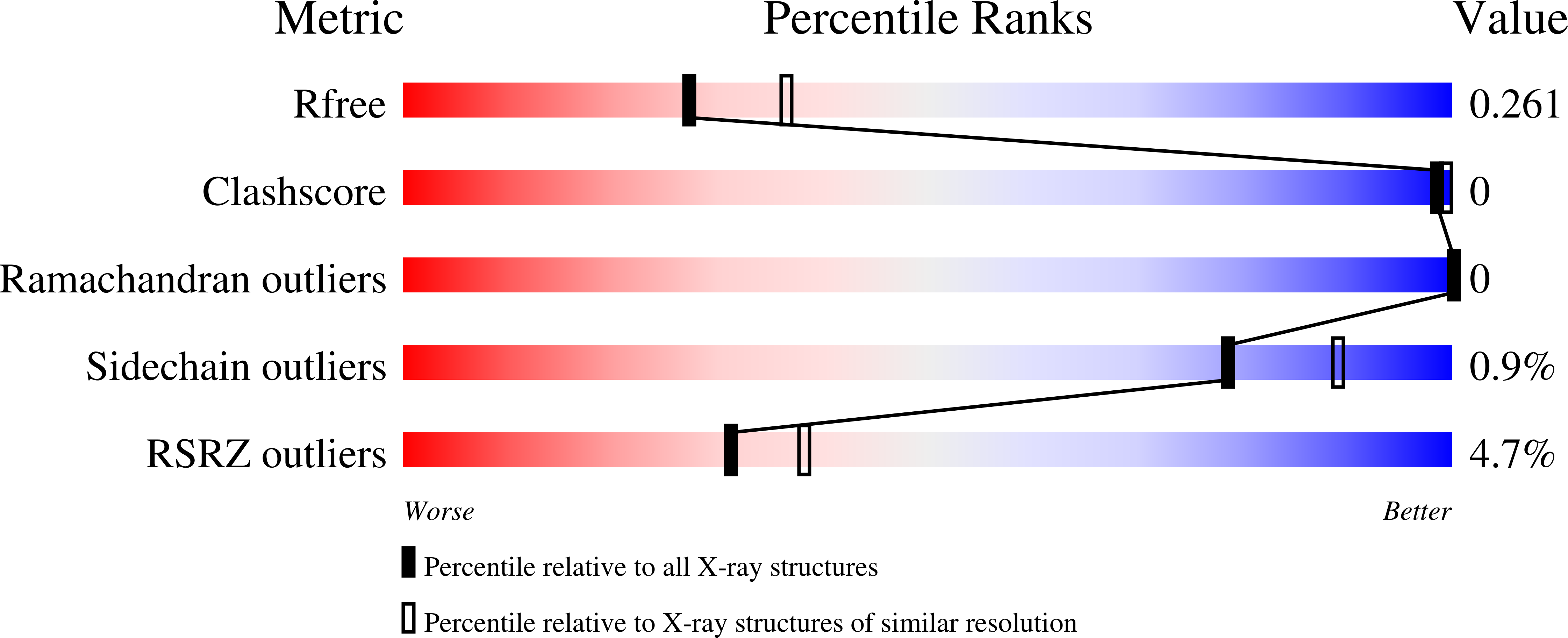
4NJL - PubMed Abstract:
A novel human coronavirus, Middle East respiratory syndrome coronavirus (MERS-CoV), has caused outbreaks of a SARS-like illness with high case fatality rate. The reports of its person-to-person transmission through close contacts have raised a global concern about its pandemic potential. Here we characterize the six-helix bundle fusion core structure of MERS-CoV spike protein S2 subunit by X-ray crystallography and biophysical analysis. We find that two peptides, HR1P and HR2P, spanning residues 998-1039 in HR1 and 1251-1286 in HR2 domains, respectively, can form a stable six-helix bundle fusion core structure, suggesting that MERS-CoV enters into the host cell mainly through membrane fusion mechanism. HR2P can effectively inhibit MERS-CoV replication and its spike protein-mediated cell-cell fusion. Introduction of hydrophilic residues into HR2P results in significant improvement of its stability, solubility and antiviral activity. Therefore, the HR2P analogues have good potential to be further developed into effective viral fusion inhibitors for treating MERS-CoV infection.
Organizational Affiliation:
1] Key Laboratory of Medical Molecular Virology of Ministries of Education and Health, Shanghai Medical College and Institute of Medical Microbiology, Fudan University, Shanghai 200032, China [2].