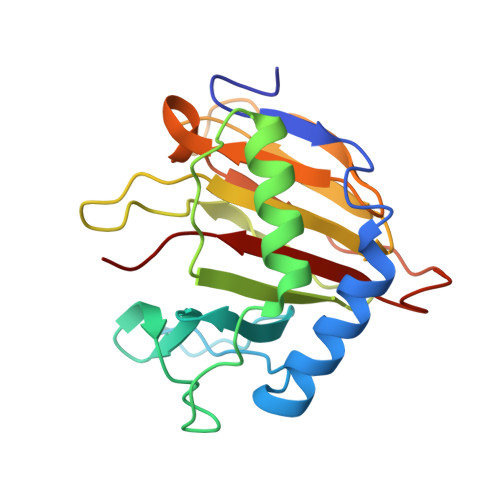
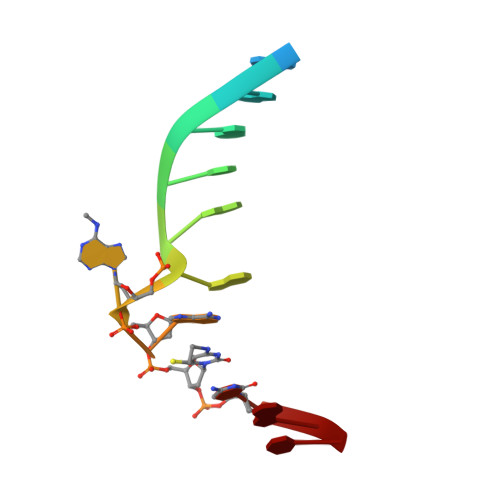
Switching Demethylation Activities between AlkB Family RNA/DNA Demethylases through Exchange of Active-Site Residues.
Zhu, C., Yi, C.(2014) Angew Chem Int Ed Engl 53: 3659-3662
- PubMed: 24596302
- DOI: https://doi.org/10.1002/anie.201310050
- Primary Citation of Related Structures:
4NID, 4NIG, 4NIH, 4NII - PubMed Abstract:
The AlkB family demethylases AlkB, FTO, and ALKBH5 recognize differentially methylated RNA/DNA substrates, which results in their distinct biological roles. Here we identify key active-site residues that contribute to their substrate specificity. Swapping such active-site residues between the demethylases leads to partially switched demethylation activities. Combined evidence from X-ray structures and enzyme kinetics suggests a role of the active-site residues in substrate recognition. Such a divergent active-site sequence may aid the design of selective inhibitors that can discriminate these homologue RNA/DNA demethylases.
Organizational Affiliation:
State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Synthetic and Functional Biomolecules Center, and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871 (China).