Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3).
Qi, S., O'Hayre, M., Gutkind, J.S., Hurley, J.H.(2014) J Biol Chem 289: 4743-4752
- PubMed: 24379409
- DOI: https://doi.org/10.1074/jbc.M113.527473
- Primary Citation of Related Structures:
4N7F, 4N7H - PubMed Abstract:
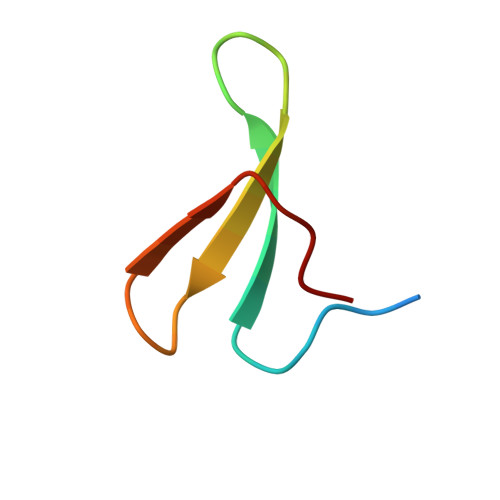
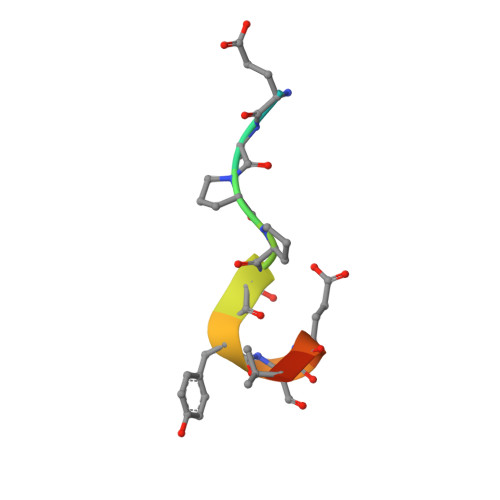
After protracted stimulation, the β2-adrenergic receptor and many other G-protein-coupled receptors are ubiquitinated and down-regulated. Arrestin-related domain-containing protein-3 (ARRDC3) has been proposed to recruit the ubiquitin ligase Nedd4 to the β2-adrenergic receptor. ARRDC3 contains two PPXY motifs that could potentially interact with any of the four WW domains of Nedd4. Here we dissect the interaction determinants. ARRDC3 PPXY-Nedd4 WW dissociation constants vary from unmeasurable to Kd = 3 μM for the third WW domain of Nedd4 binding to the first PPXY motif of ARRDC3. Structures of the uncomplexed and PPXY1-bound WW3 domain were determined at 1.1 and 1.7 Å resolution. The structures revealed conformational changes upon binding and the hydrogen bonding network in exquisite detail. Tight packing of ARRDC3 Val-352', part of a 310 helix at the C terminus of PPXY1, is important for high affinity binding to WW3. Although no single WW domain is strictly essential for the binding of Nedd4 and ARRDC3 expressed in HEK293 cells, high affinity binding of full-length ARRDC3 and Nedd4 is driven by the avid interaction of both PPXY motifs with either the WW2-WW3 or WW3-WW4 combinations, with Kd values as low as 300 nM.
Organizational Affiliation:
From the Department of Molecular and Cell Biology and California Institute for Quantitative Biosciences, University of California, Berkeley, Berkeley, California 94720 and.