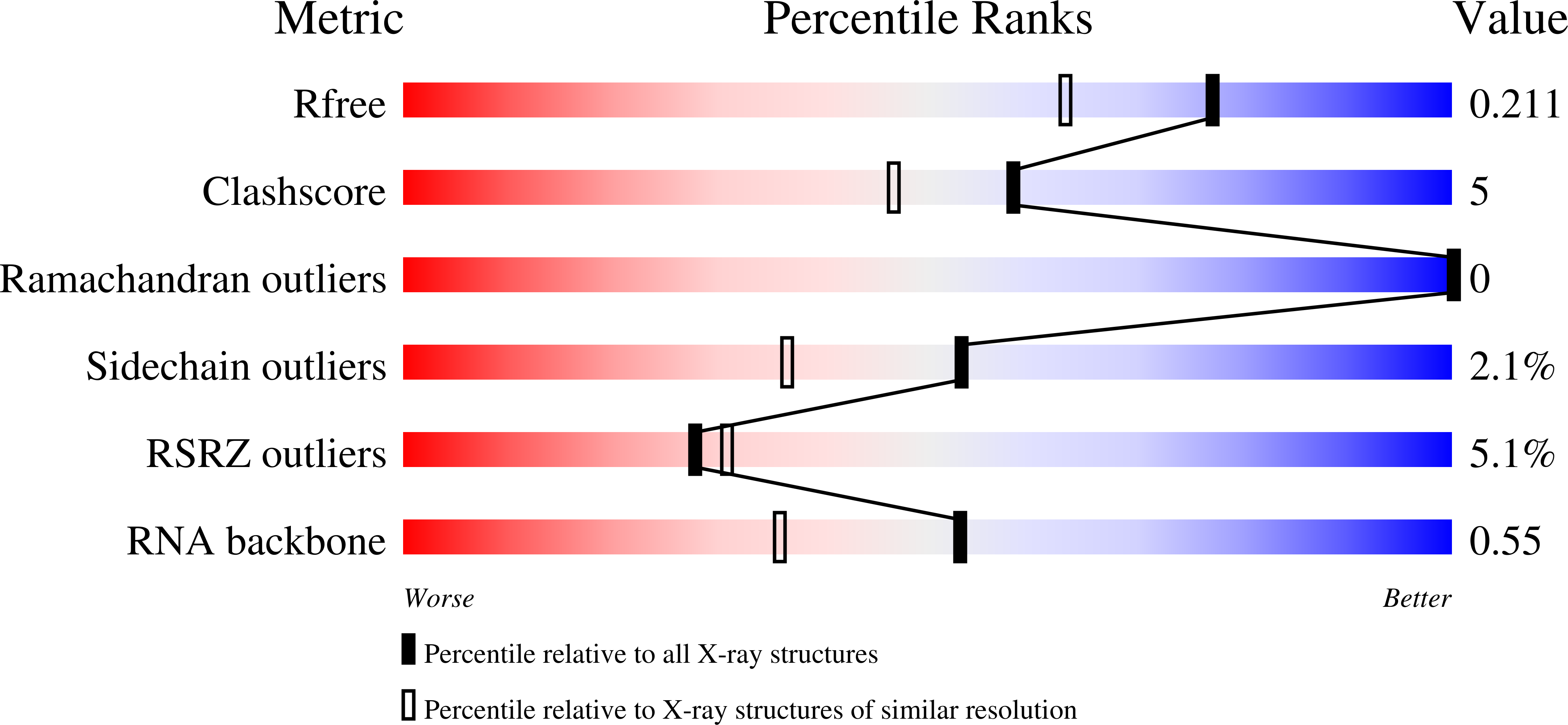
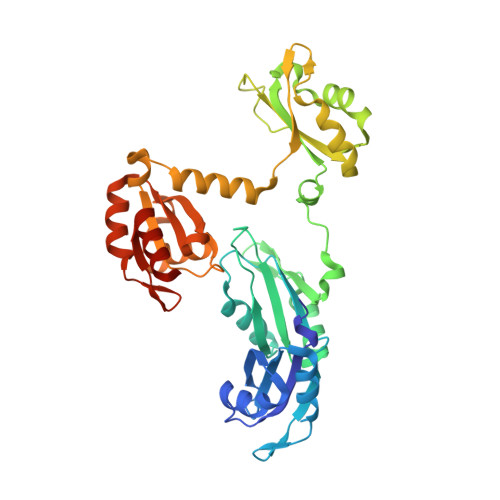
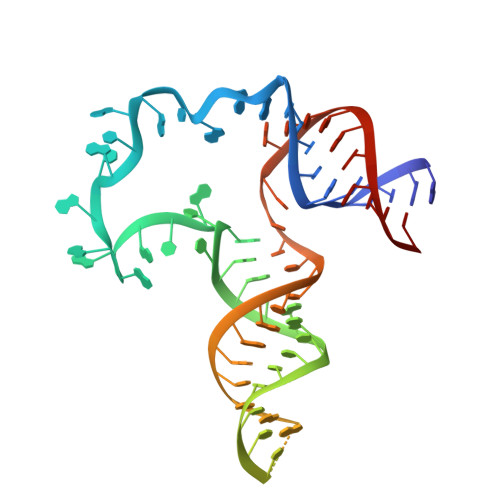
Core structure of the U6 small nuclear ribonucleoprotein at 1.7- angstrom resolution.
Montemayor, E.J., Curran, E.C., Liao, H.H., Andrews, K.L., Treba, C.N., Butcher, S.E., Brow, D.A.(2014) Nat Struct Mol Biol 21: 544-551
- PubMed: 24837192
- DOI: https://doi.org/10.1038/nsmb.2832
- Primary Citation of Related Structures:
4N0T - PubMed Abstract:
The spliceosome is a dynamic assembly of five small nuclear ribonucleoproteins (snRNPs) that removes introns from eukaryotic pre-mRNA. U6, the most conserved of the spliceosomal small nuclear RNAs (snRNAs), participates directly in catalysis. Here, we report the crystal structure of the Saccharomyces cerevisiae U6 snRNP core containing most of the U6 snRNA and all four RRM domains of the Prp24 protein. It reveals a unique interlocked RNP architecture that sequesters the 5' splice site-binding bases of U6 snRNA. RRMs 1, 2 and 4 of Prp24 form an electropositive groove that binds double-stranded RNA and may nucleate annealing of U4 and U6 snRNAs. Substitutions in Prp24 that suppress a mutation in U6 localize to direct RNA-protein contacts. Our results provide the most comprehensive view to date of a multi-RRM protein bound to RNA and reveal striking coevolution of protein and RNA structure.
Organizational Affiliation:
1] Department of Biomolecular Chemistry, University of Wisconsin-Madison, Madison, Wisconsin, USA. [2] Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin, USA.