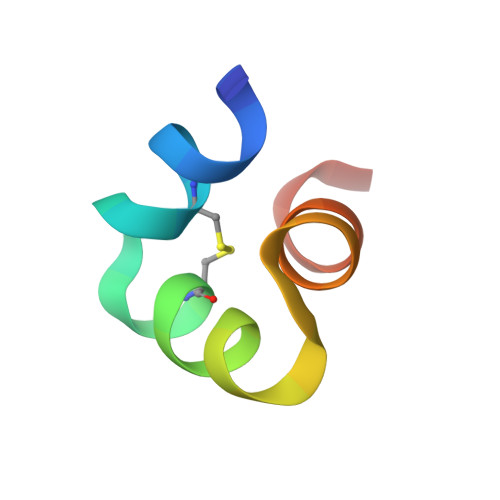X-ray crystal structure of bovine 3 Glu-osteocalcin.
Malashkevich, V.N., Almo, S.C., Dowd, T.L.(2013) Biochemistry 52: 8387-8392
- PubMed: 24138653
- DOI: https://doi.org/10.1021/bi4010254
- Primary Citation of Related Structures:
4MZZ - PubMed Abstract:
The 3 Glu form of osteocalcin (3 Glu-OCN) is increased in serum during low vitamin K intake or oral anticoagulant use (warfarin). Previous reports using circular dichroism show it is less structured than 3 Gla Ca²⁺-osteocalcin and does not bind strongly to bone mineral. Recent studies have suggested a role for 3 Glu-OCN as a potential regulator of glucose metabolism. A G-protein-coupled receptor, GPRC6a, found in the pancreas and testes was identified as the putative osteocalcin receptor. The purpose of this study is to determine the high-resolution structure of bovine 3 Glu-OCN, using X-ray crystallography, to understand molecular interactions with mineral and the GPRC6a receptor. Diffraction quality crystals of thermally decarboxylated bovine osteocalcin were grown, and the crystal structure was determined to 1.88 Å resolution. The final refined structure contained residues 17-47 and, like 3 Gla Ca²⁺-OCN, consisted of three α-helices surrounding a hydrophobic core, a C23-C29 disulfide bond between two of the helices, and no bound Ca²⁺. Thus, the helical structure of 3 Glu-OCN is Ca²⁺-independent but similar to that of 3 Gla Ca²⁺-OCN. A reduced level of mineral binding could result from a lower number of Ca²⁺ coordinating ligands on 3 Glu-OCN. The structure suggests the GPRC6a receptor may respond to helical osteocalcin and will aid in providing molecular mechanistic insight into the role of 3 Glu-OCN in glucose homeostasis.
Organizational Affiliation:
Department of Biochemistry, Albert Einstein College of Medicine , Bronx, New York 10461, United States.