Crystal Structure of the putative tail fiber protein gp53 from the Acinetobacter baumannii bacteriophage AP22
Sycheva, L.V., Shneider, M.M., Popova, A.V., Ziganshin, R.K., Volozhantsev, N., Miroshnikov, K.A., Leiman, P.G.(2019) Biorxiv
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2019) Biorxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative tail fiber protein | 164 | Acinetobacter phage AP22 | Mutation(s): 0 Gene Names: ORF53 | 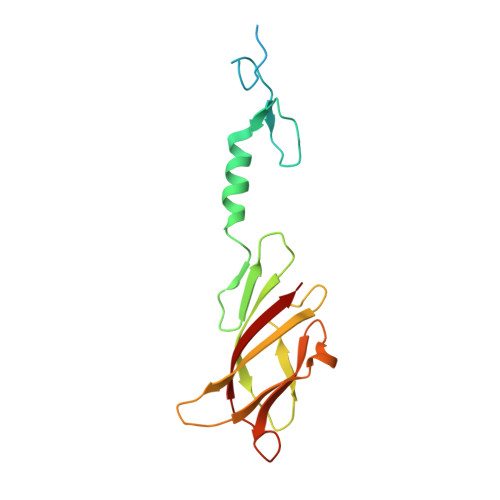 | |
UniProt | |||||
Find proteins for I2GUG0 (Acinetobacter phage AP22) Explore I2GUG0 Go to UniProtKB: I2GUG0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | I2GUG0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | B [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| SO3 Query on SO3 | BA [auth A] | SULFITE ION O3 S LSNNMFCWUKXFEE-UHFFFAOYSA-L |  | ||
| BR Query on BR | C [auth A] D [auth A] E [auth A] F [auth A] G [auth A] | BROMIDE ION Br CPELXLSAUQHCOX-UHFFFAOYSA-M |  | ||
| EDO Query on EDO | AA [auth A], Z [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| NA Query on NA | W [auth A], X [auth A], Y [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 51.2 | α = 90 |
| b = 51.2 | β = 90 |
| c = 302.03 | γ = 120 |
| Software Name | Purpose |
|---|---|
| MAR345 | data collection |
| SHELX | model building |
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| SHELX | phasing |