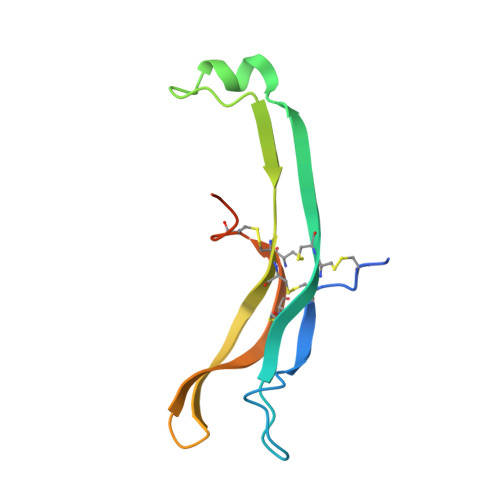
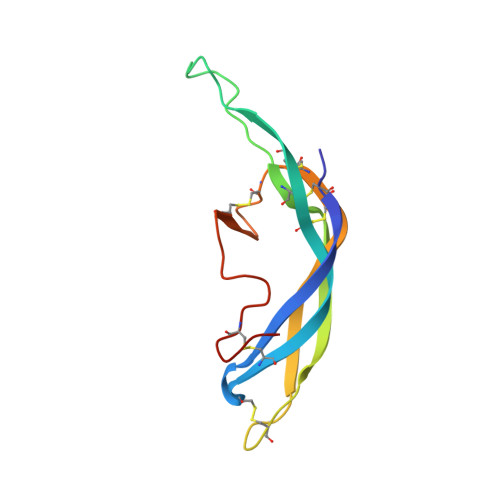
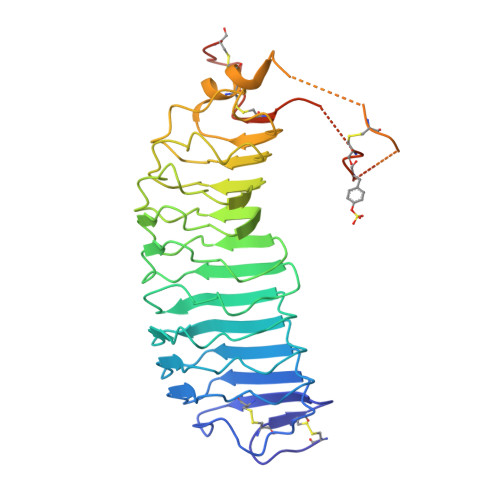
Evidence for Follicle-stimulating Hormone Receptor as a Functional Trimer.
Jiang, X., Fischer, D., Chen, X., McKenna, S.D., Liu, H., Sriraman, V., Yu, H.N., Goutopoulos, A., Arkinstall, S., He, X.(2014) J Biol Chem 289: 14273-14282
- PubMed: 24692546
- DOI: https://doi.org/10.1074/jbc.M114.549592
- Primary Citation of Related Structures:
4MQW - PubMed Abstract:
Follicle-stimulating hormone receptor (FSHR), a G-protein coupled receptor, is an important drug target in the development of novel therapeutics for reproductive indications. The FSHR extracellular domains were observed in the crystal structure as a trimer, which enabled us to propose a novel model for the receptor activation mechanism. The model predicts that FSHR binds Asnα(52)-deglycosylated FSH at a 3-fold higher capacity than fully glycosylated FSH. It also predicts that, upon dissociation of the FSHR trimer into monomers, the binding of glycosylated FSH, but not deglycosylated FSH, would increase 3-fold, and that the dissociated monomers would in turn enhance FSHR binding and signaling activities by 3-fold. This study presents evidence confirming these predictions and provides crystallographic and mutagenesis data supporting the proposed model. The model also provides a mechanistic explanation to the agonist and antagonist activities of thyroid-stimulating hormone receptor autoantibodies. We conclude that FSHR exists as a functional trimer.
Organizational Affiliation:
From the EMD Serono Research and Development Institute, Billerica, Massachusetts 01821 and xuliang.jiang@emdserono.com.