Structural insight into DNA binding and oligomerization of the multifunctional Cox protein of bacteriophage P2.
Berntsson, R.P., Odegrip, R., Sehlen, W., Skaar, K., Svensson, L.M., Massad, T., Hogbom, M., Haggard-Ljungquist, E., Stenmark, P.(2014) Nucleic Acids Res 42: 2725-2735
- PubMed: 24259428
- DOI: https://doi.org/10.1093/nar/gkt1119
- Primary Citation of Related Structures:
4LHF - PubMed Abstract:
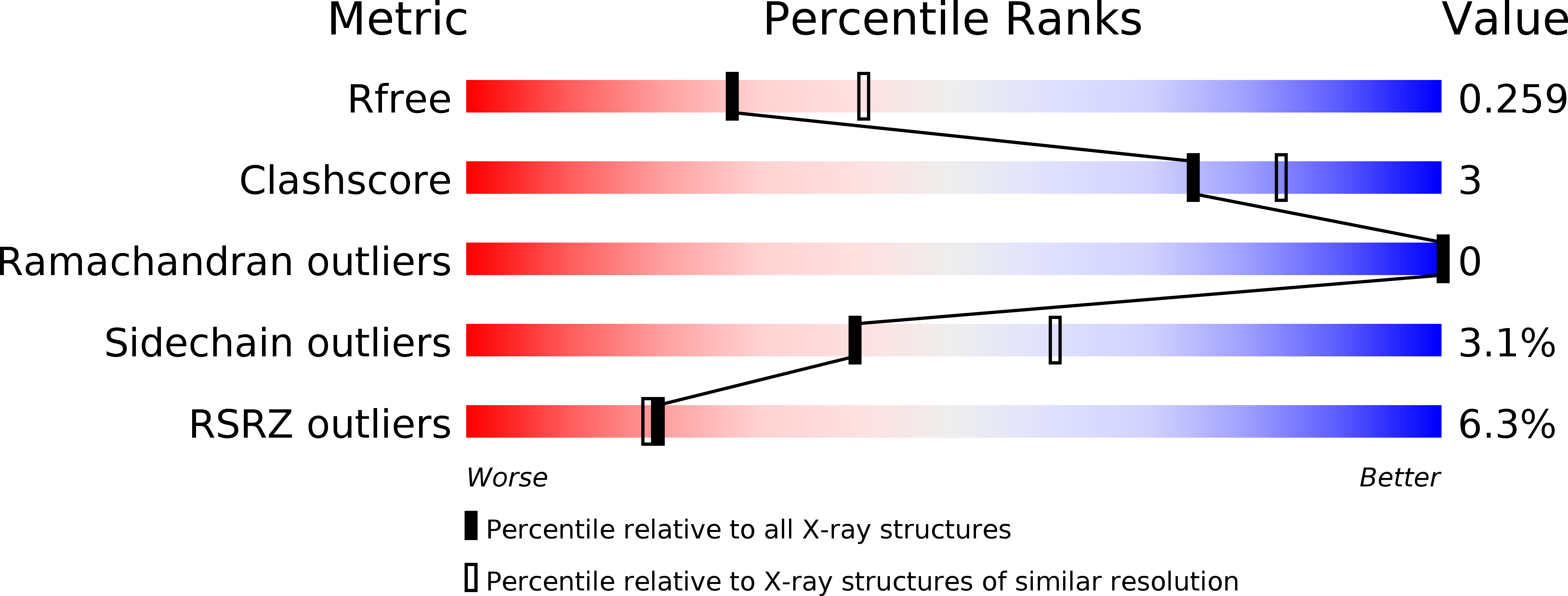
The Cox protein from bacteriophage P2 is a small multifunctional DNA-binding protein. It is involved in site-specific recombination leading to P2 prophage excision and functions as a transcriptional repressor of the P2 Pc promoter. Furthermore, it transcriptionally activates the unrelated, defective prophage P4 that depends on phage P2 late gene products for lytic growth. In this article, we have investigated the structural determinants to understand how P2 Cox performs these different functions. We have solved the structure of P2 Cox to 2.4 Å resolution. Interestingly, P2 Cox crystallized in a continuous oligomeric spiral with its DNA-binding helix and wing positioned outwards. The extended C-terminal part of P2 Cox is largely responsible for the oligomerization in the structure. The spacing between the repeating DNA-binding elements along the helical P2 Cox filament is consistent with DNA binding along the filament. Functional analyses of alanine mutants in P2 Cox argue for the importance of key residues for protein function. We here present the first structure from the Cox protein family and, together with previous biochemical observations, propose that P2 Cox achieves its various functions by specific binding of DNA while wrapping the DNA around its helical oligomer.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Arrhenius Laboratories for Natural Sciences, Stockholm University, SE-10691 Stockholm, Sweden and Department of Molecular Biosciences, The Wenner-Gren Institute, Arrhenius Laboratories for Natural Sciences, Stockholm University, SE-10691 Stockholm, Sweden.