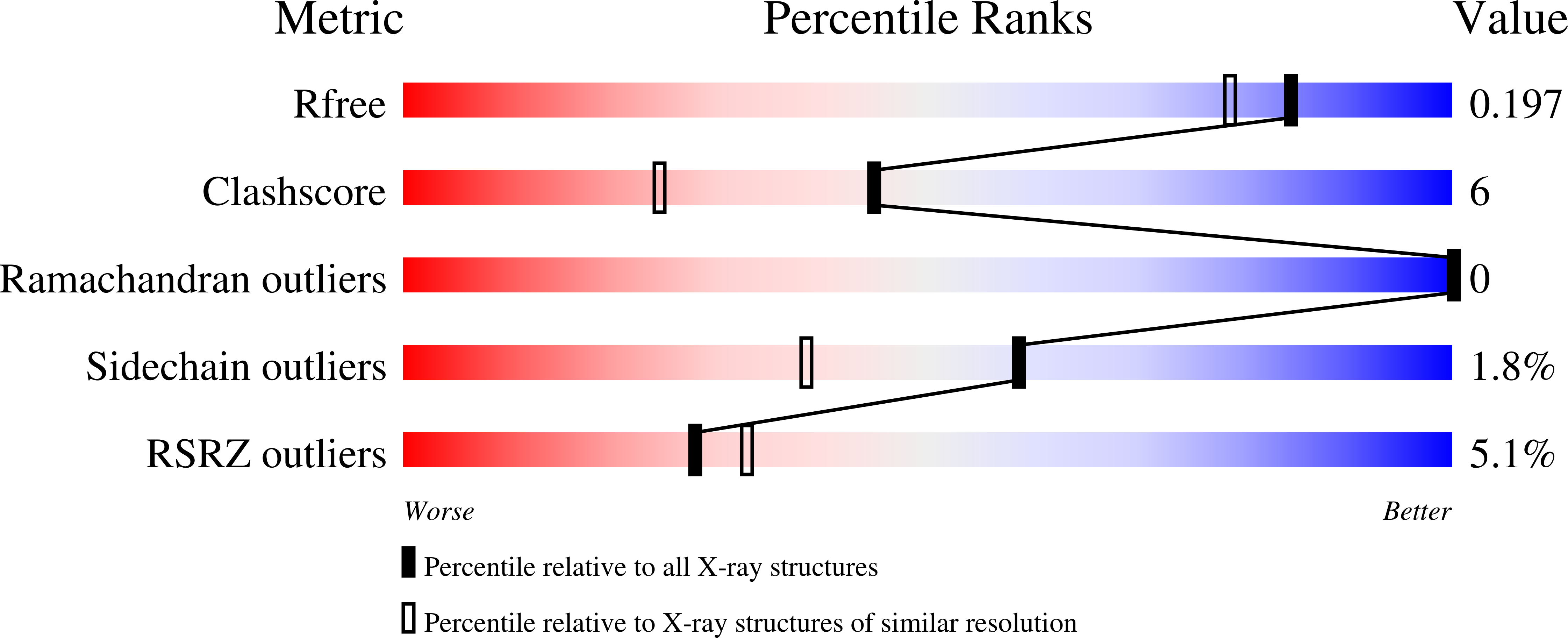
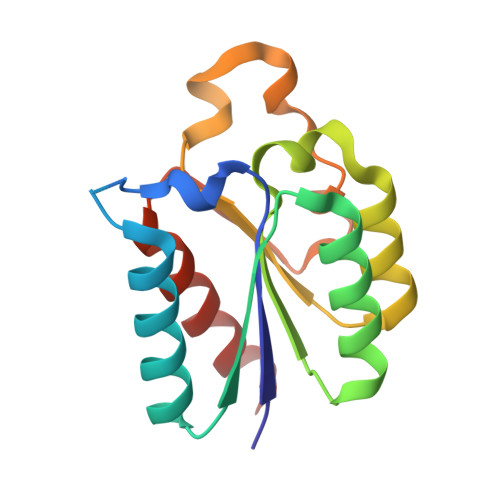
Structure of type II dehydroquinase from Pseudomonas aeruginosa.
Reiling, S., Kelleher, A., Matsumoto, M.M., Robinson, G., Asojo, O.A.(2014) Acta Crystallogr F Struct Biol Commun 70: 1485-1491
- PubMed: 25372814
- DOI: https://doi.org/10.1107/S2053230X14020214
- Primary Citation of Related Structures:
4L8L - PubMed Abstract:
Pseudomonas aeruginosa causes opportunistic infections and is resistant to most antibiotics. Ongoing efforts to generate much-needed new antibiotics include targeting enzymes that are vital for P. aeruginosa but are absent in mammals. One such enzyme, type II dehydroquinase (DHQase), catalyzes the interconversion of 3-dehydroquinate and 3-dehydroshikimate, a necessary step in the shikimate pathway. This step is vital for the proper synthesis of phenylalanine, tryptophan, tyrosine and other aromatic metabolites. The recombinant expression, purification and crystal structure of catalytically active DHQase from P. aeruginosa (PaDHQase) are presented. Cubic crystals belonging to space group F23, with unit-cell parameters a=b=c=125.39 Å, were obtained by vapor diffusion in sitting drops and the structure was refined to an R factor of 16% at 1.74 Å resolution. PaDHQase is a prototypical type II DHQase with the classical flavodoxin-like α/β topology.
Organizational Affiliation:
Toxicology Department, School of Public Health University, University of Nebraska Medical Center, Omaha, NE 68198, USA.