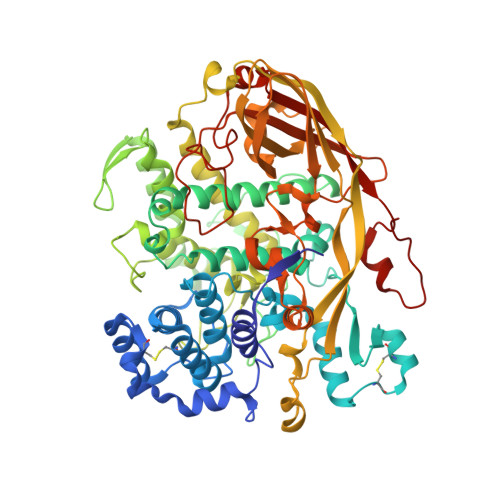
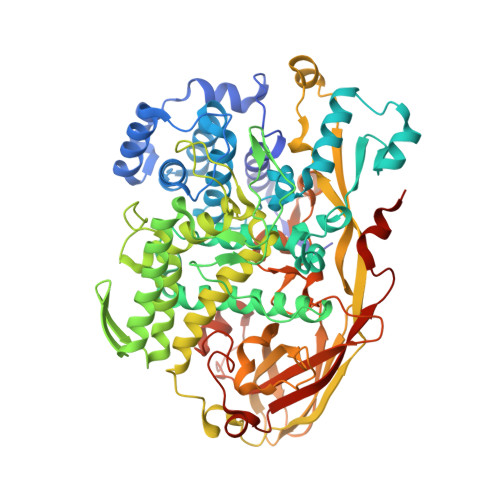
Crystallographic identification of an unexpected protein complex in silkworm haemolymph.
Pietrzyk, A.J., Bujacz, A., Mueller-Dieckmann, J., ochynska, M., Jaskolski, M., Bujacz, G.(2013) Acta Crystallogr D Biol Crystallogr 69: 2353-2364
- PubMed: 24311577
- DOI: https://doi.org/10.1107/S0907444913021823
- Primary Citation of Related Structures:
4L37 - PubMed Abstract:
The first crystal structure of a complex formed by two storage proteins, SP2 and SP3, isolated from their natural source, mulberry silkworm (Bombyx mori L.) haemolymph, has been determined. The structure was solved by molecular replacement using arylphorin, a protein rich in aromatic amino-acid residues, from oak silkworm as the initial model. The quality of the electron-density maps obtained from the X-ray diffraction experiment allowed the authors to detect that the investigated crystal structure was composed of two different arylphorins: SP2 and SP3. This discovery was confirmed by N-terminal sequencing. SP2 has been extensively studied previously, whereas only a few reports on SP3 are available. However, to date no structural studies have been reported for these proteins. These studies revealed that SP2 and SP3 exist in the silkworm body as a heterohexamer formed by one SP2 trimer and one SP3 trimer. The overall fold, consisting of three haemocyanin-like subdomains, of SP2 and SP3 is similar. Both proteins contain a conserved N-glycosylation motif in their structures.
Organizational Affiliation:
Center for Biocrystallographic Research, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14, 61-704 Poznan, Poland.