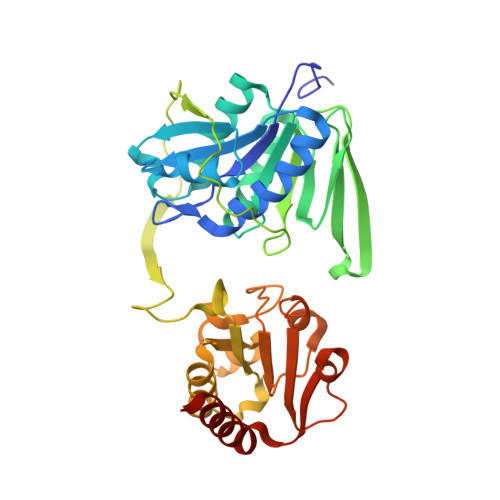
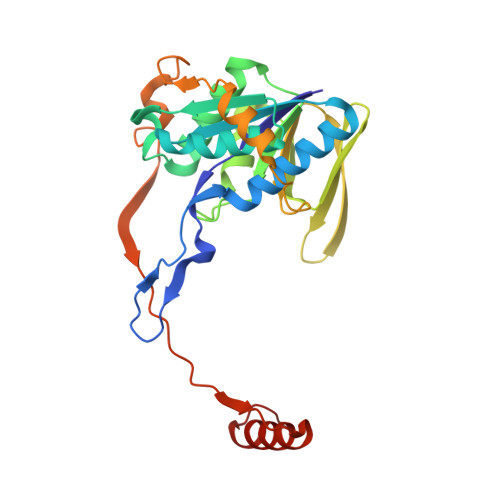
Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
Chowdhury, N.P., Mowafy, A.M., Demmer, J.K., Upadhyay, V., Koelzer, S., Jayamani, E., Kahnt, J., Hornung, M., Demmer, U., Ermler, U., Buckel, W.(2014) J Biol Chem 289: 5145-5157
- PubMed: 24379410
- DOI: https://doi.org/10.1074/jbc.M113.521013
- Primary Citation of Related Structures:
4KPU, 4L1F, 4L2I - PubMed Abstract:
Electron bifurcation is a fundamental strategy of energy coupling originally discovered in the Q-cycle of many organisms. Recently a flavin-based electron bifurcation has been detected in anaerobes, first in clostridia and later in acetogens and methanogens. It enables anaerobic bacteria and archaea to reduce the low-potential [4Fe-4S] clusters of ferredoxin, which increases the efficiency of the substrate level and electron transport phosphorylations. Here we characterize the bifurcating electron transferring flavoprotein (EtfAf) and butyryl-CoA dehydrogenase (BcdAf) of Acidaminococcus fermentans, which couple the exergonic reduction of crotonyl-CoA to butyryl-CoA to the endergonic reduction of ferredoxin both with NADH. EtfAf contains one FAD (α-FAD) in subunit α and a second FAD (β-FAD) in subunit β. The distance between the two isoalloxazine rings is 18 Å. The EtfAf-NAD(+) complex structure revealed β-FAD as acceptor of the hydride of NADH. The formed β-FADH(-) is considered as the bifurcating electron donor. As a result of a domain movement, α-FAD is able to approach β-FADH(-) by about 4 Å and to take up one electron yielding a stable anionic semiquinone, α-FAD, which donates this electron further to Dh-FAD of BcdAf after a second domain movement. The remaining non-stabilized neutral semiquinone, β-FADH(•), immediately reduces ferredoxin. Repetition of this process affords a second reduced ferredoxin and Dh-FADH(-) that converts crotonyl-CoA to butyryl-CoA.
Organizational Affiliation:
From the Laboratorium für Mikrobiologie, Fachbereich Biologie and SYNMIKRO, Philipps-Universität, 35032 Marburg, Germany.