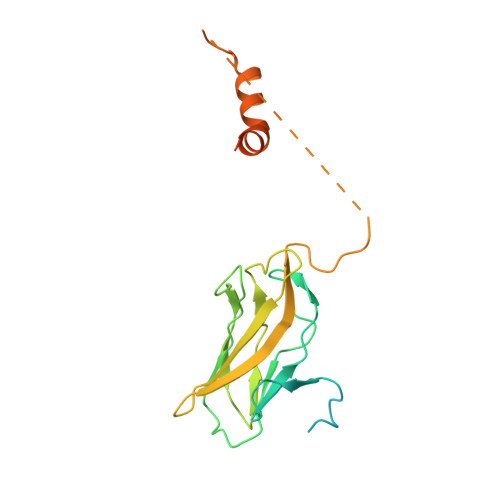
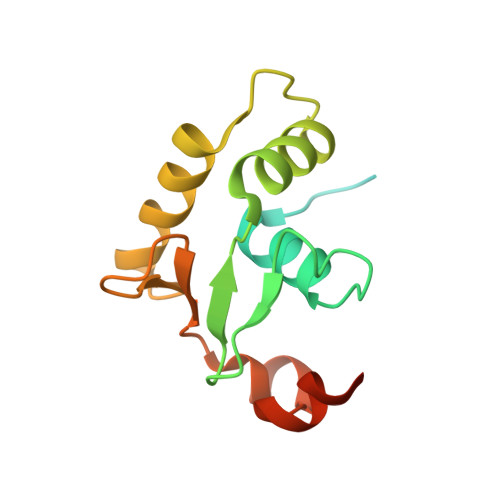
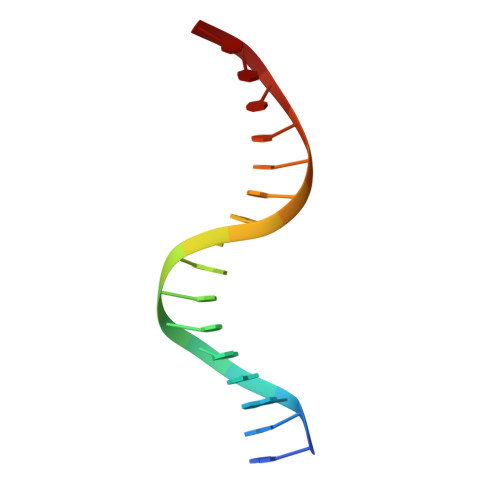
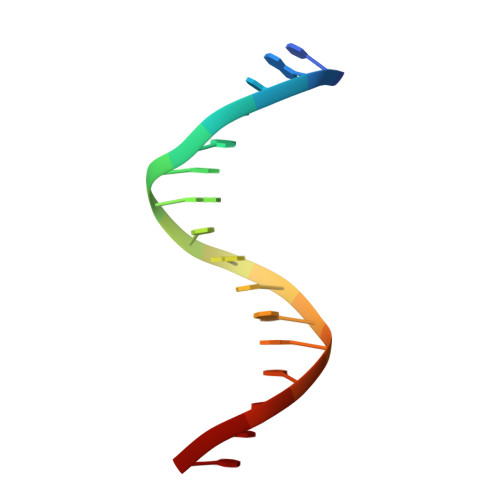
Structural basis of Ets1 activation by Runx1.
Shrivastava, T., Mino, K., Babayeva, N.D., Baranovskaya, O.I., Rizzino, A., Tahirov, T.H.(2014) Leukemia 28: 2040-2048
- PubMed: 24646888
- DOI: https://doi.org/10.1038/leu.2014.111
- Primary Citation of Related Structures:
4L0Y, 4L0Z, 4L18 - PubMed Abstract:
Runx1 is required for definitive hematopoiesis and is well known for its frequent chromosomal translocations and point mutations in leukemia. Runx1 regulates a variety of genes via Ets1 activation on an Ets1•Runx1 composite DNA sequence. The structural basis of such regulation remains unresolved. To address this problem, we determined the crystal structure of the ternary complex containing Runx1(1-242) and Ets1(296-441) bound to T-cell receptor alpha (TCRα) enhancer DNA. In the crystal, an Ets1-interacting domain of Runx1 is bound to the Ets1 DNA-binding domain and displaced an entire autoinhibitory module of Ets1, revealing a novel mechanism of Ets1 activation. The DNA-binding and transcriptional studies with a variety of structure-guided Runx1 mutants confirmed a critical role of direct Ets1•Runx1 interaction in Ets1 activation. More importantly, the discovered mechanism provides a plausible explanation for how the Ets1•Runx1 interaction effectively activates not only a wild-type Ets1, but also a highly inhibited phosphorylated form of Ets1.
Organizational Affiliation:
Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, NE, USA.