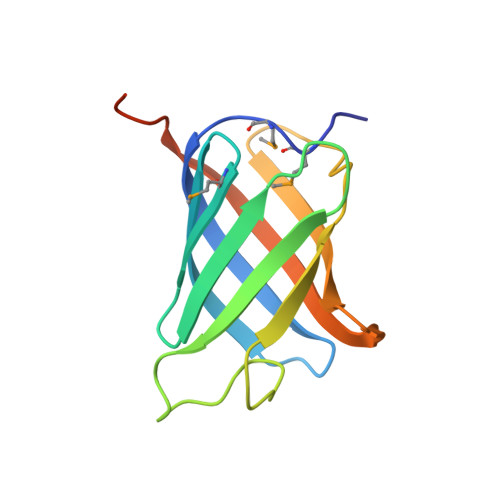
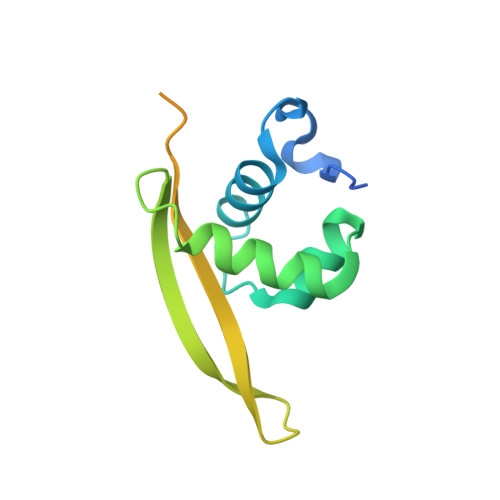
Structure of GrlR-GrlA complex that prevents GrlA activation of virulence genes
Padavannil, A., Jobichen, C., Mills, E., Velazquez-Campoy, A., Li, M., Leung, K.Y., Mok, Y.K., Rosenshine, I., Sivaraman, J.(2013) Nat Commun 4: 2546-2546
- PubMed: 24092262
- DOI: https://doi.org/10.1038/ncomms3546
- Primary Citation of Related Structures:
4KT5 - PubMed Abstract:
The locus of enterocyte effacement (LEE) is essential for virulence of enterohaemorrhagic Escherichia coli (EHEC) and enteropathogenic E. coli (EPEC). The 41 genes of the LEE encode type III secretion system proteins and three associated regulators: Ler, GrlA and GrlR. Ler is a positive regulator for most of the LEE operons, including grlRA. GrlA controls the expression of ler, ehxCABD and flhDC operons. GrlR binds to GrlA and suppresses its function. Here we report the crystal structure of GrlR-GrlAΔ (aa 1-106) complex (2:1) and its functional characterization. We show that GrlR interacts with the Helix-Turn-Helix motif of GrlA. Moreover, GrlA binds to the promoter DNA fragments of ler, ehxCABD and flhDC, and GrlR outcompetes with these promoter DNA sequences for the Helix-Turn-Helix motif of GrlA. These findings provide mechanistic insight into a regulatory module for the virulence of EPEC and EHEC, two important pathogens that cause devastating diseases.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543, Singapore.