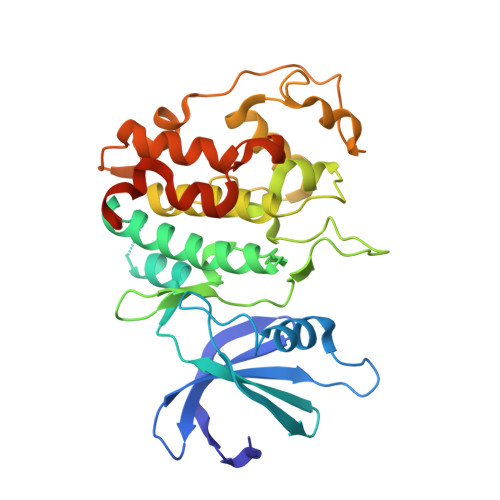
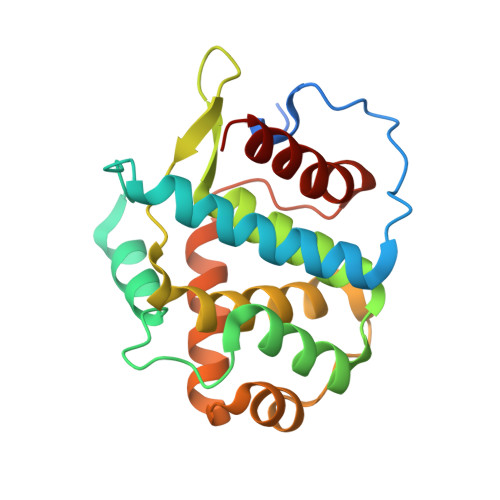
New Structural Insights into Phosphorylation-free Mechanism for Full Cyclin-dependent Kinase (CDK)-Cyclin Activity and Substrate Recognition.
Zheng, F., Quiocho, F.A.(2013) J Biol Chem 288: 30682-30692
- PubMed: 24022486
- DOI: https://doi.org/10.1074/jbc.M113.502773
- Primary Citation of Related Structures:
4KRC, 4KRD - PubMed Abstract:
Pho85 is a versatile cyclin-dependent kinase (CDK) found in budding yeast that regulates a myriad of eukaryotic cellular functions in concert with 10 cyclins (called Pcls). Unlike cell cycle CDKs that require phosphorylation of a serine/threonine residue by a CDK-activating kinase (CAK) for full activation, Pho85 requires no phosphorylation despite the presence of an equivalent residue. The Pho85-Pcl10 complex is a key regulator of glycogen metabolism by phosphorylating the substrate Gsy2, the predominant, nutritionally regulated form of glycogen synthase. Here we report the crystal structures of Pho85-Pcl10 and its complex with the ATP analog, ATPγS. The structure solidified the mechanism for bypassing CDK phosphorylation to achieve full catalytic activity. An aspartate residue, invariant in all Pcls, acts as a surrogate for the phosphoryl adduct of the phosphorylated, fully activated CDK2, the prototypic cell cycle CDK, complexed with cyclin A. Unlike the canonical recognition motif, SPX(K/R), of phosphorylation sites of substrates of several cell cycle CDKs, the motif in the Gys2 substrate of Pho85-Pcl10 is SPXX. CDK5, an important signal transducer in neural development and the closest known functional homolog of Pho85, does not require phosphorylation either, and we found that in its crystal structure complexed with p25 cyclin a water/hydroxide molecule remarkably plays a similar role to the phosphoryl or aspartate group. Comparison between Pho85-Pcl10, phosphorylated CDK2-cyclin A, and CDK5-p25 complexes reveals the convergent structural characteristics necessary for full kinase activity and the variations in the substrate recognition mechanism.
Organizational Affiliation:
From the Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, Texas 77030.