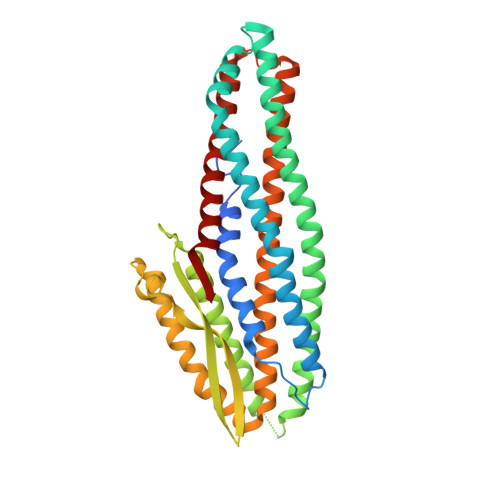
Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Ganash, M., Phung, D., Sedelnikova, S.E., Lindback, T., Granum, P.E., Artymiuk, P.J.(2013) PLoS One 8: e74748-e74748
- PubMed: 24040335
- DOI: https://doi.org/10.1371/journal.pone.0074748
- Primary Citation of Related Structures:
4K1P - PubMed Abstract:
The structure of NheA, a component of the Bacillus cereus Nhe tripartite toxin, has been solved at 2.05 Å resolution using selenomethionine multiple-wavelength anomalous dispersion (MAD). The structure shows it to have a fold that is similar to the Bacillus cereus Hbl-B and E. coli ClyA toxins, and it is therefore a member of the ClyA superfamily of α-helical pore forming toxins (α-PFTs), although its head domain is significantly enlarged compared with those of ClyA or Hbl-B. The hydrophobic β-hairpin structure that is a characteristic of these toxins is replaced by an amphipathic β-hairpin connected to the main structure via a β-latch that is reminiscent of a similar structure in the β-PFT Staphylococcus aureus α-hemolysin. Taken together these results suggest that, although it is a member of an archetypal α-PFT family of toxins, NheA may be capable of forming a β rather than an α pore.
Organizational Affiliation:
The Krebs Institute, Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, United Kingdom.