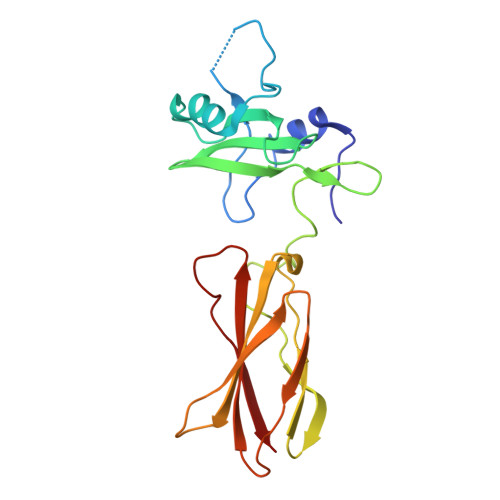
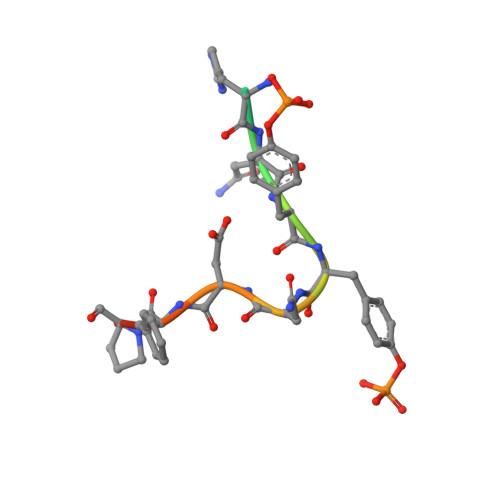
Directed network wiring identifies a key protein interaction in embryonic stem cell differentiation.
Yasui, N., Findlay, G.M., Gish, G.D., Hsiung, M.S., Huang, J., Tucholska, M., Taylor, L., Smith, L., Boldridge, W.C., Koide, A., Pawson, T., Koide, S.(2014) Mol Cell 54: 1034-1041
- PubMed: 24910098
- DOI: https://doi.org/10.1016/j.molcel.2014.05.002
- Primary Citation of Related Structures:
4JMG, 4JMH - PubMed Abstract:
Cell signaling depends on dynamic protein-protein interaction (PPI) networks, often assembled through modular domains each interacting with multiple peptide motifs. This complexity raises a conceptual challenge, namely to define whether a particular cellular response requires assembly of the complete PPI network of interest or can be driven by a specific interaction. To address this issue, we designed variants of the Grb2 SH2 domain ("pY-clamps") whose specificity is highly biased toward a single phosphotyrosine (pY) motif among many potential pYXNX Grb2-binding sites. Surprisingly, directing Grb2 predominantly to a single pY site of the Ptpn11/Shp2 phosphatase, but not other sites tested, was sufficient for differentiation of the essential primitive endoderm lineage from embryonic stem cells. Our data suggest that discrete connections within complex PPI networks can underpin regulation of particular biological events. We propose that this directed wiring approach will be of general utility in functionally annotating specific PPIs.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA.