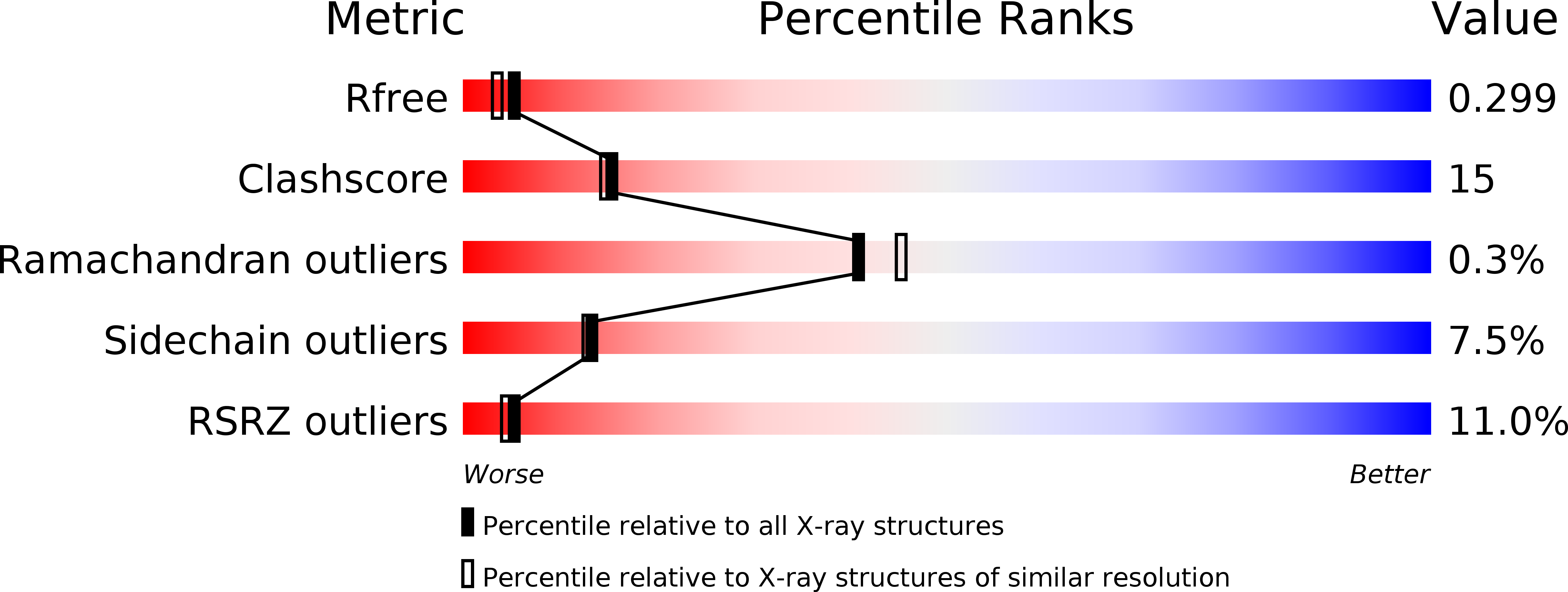
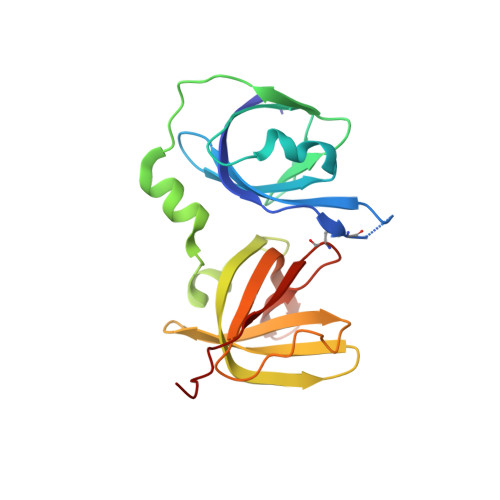
Structural basis for termination of AIM2-mediated signaling by p202
Ru, H., Ni, X., Zhao, L., Crowley, C., Ding, W., Hung, L.-W., Shaw, N., Cheng, G., Liu, Z.-J.(2013) Cell Res 23: 855-858
- PubMed: 23567559
- DOI: https://doi.org/10.1038/cr.2013.52
- Primary Citation of Related Structures:
4JBJ, 4JBK, 4JBM