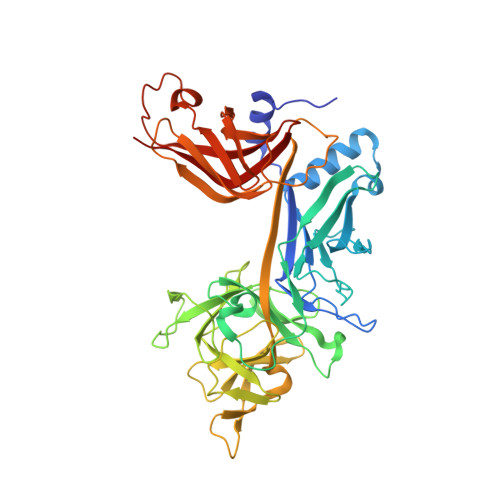
Structure and Function of the CSL-KyoT2 Corepressor Complex: A Negative Regulator of Notch Signaling.
Collins, K.J., Yuan, Z., Kovall, R.A.(2014) Structure 22: 70-81
- PubMed: 24290140
- DOI: https://doi.org/10.1016/j.str.2013.10.010
- Primary Citation of Related Structures:
4J2X - PubMed Abstract:
Notch refers to a highly conserved cell-to-cell signaling pathway with essential roles in embryonic development and tissue maintenance. Dysfunctional signaling causes human disease, highlighting the importance of pathway regulation. Notch signaling ultimately results in the activation of target genes, which is regulated by the nuclear effector CSL (CBF-1/RBP-J, Su(H), Lag-1). CSL dually functions as an activator and a repressor of transcription through differential interactions with coactivator or corepressor proteins, respectively. Although the structures of CSL-coactivator complexes have been determined, the structures of CSL-corepressor complexes are unknown. Here, using a combination of structural, biophysical, and cellular approaches, we characterize the structure and function of CSL in complex with the corepressor KyoT2. Collectively, our studies provide molecular insights into how KyoT2 binds CSL with high affinity and competes with coactivators, such as Notch, for binding CSL. These studies are important for understanding how CSL functions as both an activator and a repressor of transcription of Notch target genes.
Organizational Affiliation:
Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati, Cincinnati, OH 45267 USA.