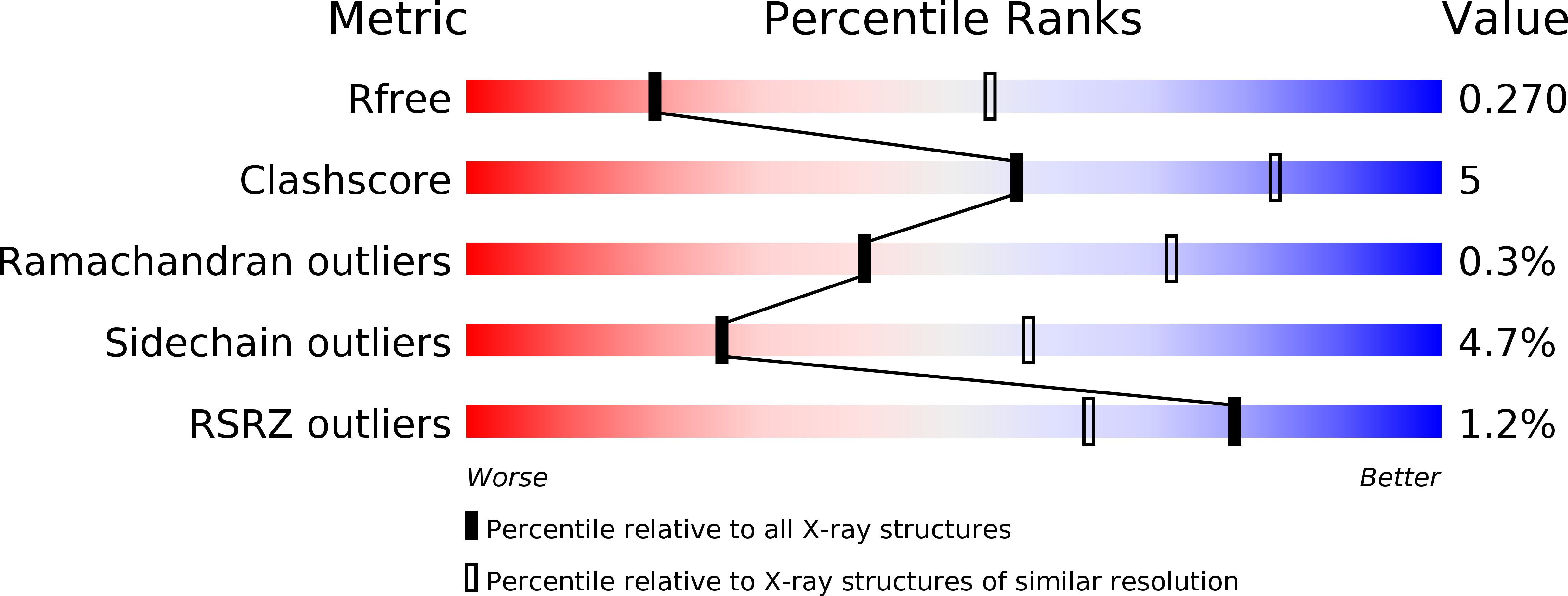
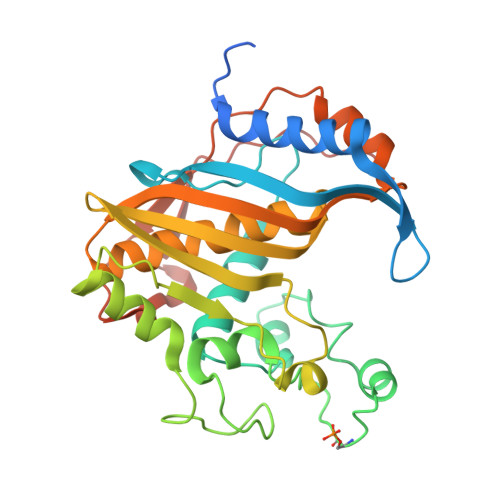
Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Wilk, P., Jarmua, A., Ruman, T., Banaszak, K., Rypniewski, W., Ciesla, J., Dowiercia, A., Rode, W.(2013) Bioorg Chem 52C: 44-49
- PubMed: 24321279
- DOI: https://doi.org/10.1016/j.bioorg.2013.11.003
- Primary Citation of Related Structures:
4ISW - PubMed Abstract:
Crystal structure is presented of the binary complex between potassium phosphoramidate-phosphorylated recombinant C. elegans thymidylate synthase and dUMP. On each monomer a single phosphoserine residue (Ser127) was identified, instead of expected phosphohistidine. As (31)P NMR studies of both the phosphorylated protein and of potassium phosphoramidate potential to phosphorylate different amino acids point to histidine as the only possible site of the modification, thermodynamically favored intermolecular phosphotransfer from histidine to serine is suggested.
Organizational Affiliation:
Nencki Institute of Experimental Biology, Polish Academy of Sciences, Warszawa, Poland.