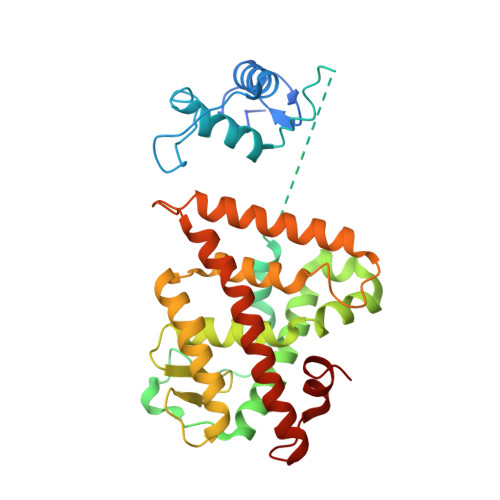
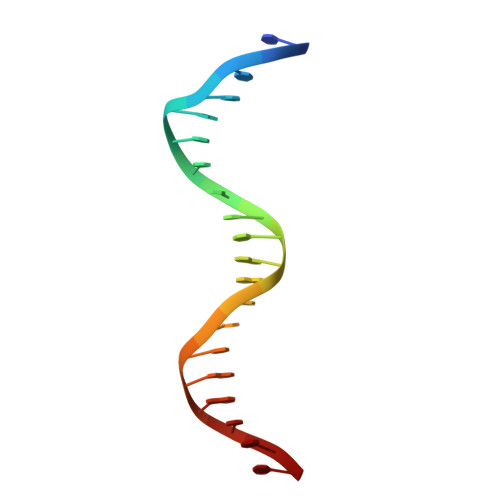
Multidomain integration in the structure of the HNF-4 alpha nuclear receptor complex.
Chandra, V., Huang, P., Potluri, N., Wu, D., Kim, Y., Rastinejad, F.(2013) Nature 495: 394-398
- PubMed: 23485969
- DOI: https://doi.org/10.1038/nature11966
- Primary Citation of Related Structures:
4IQR - PubMed Abstract:
The hepatocyte nuclear factor 4α (HNF-4α; also known as NR2A1) is a member of the nuclear receptor (NR) family of transcription factors, which have conserved DNA-binding domains and ligand-binding domains. HNF-4α is the most abundant DNA-binding protein in the liver, where some 40% of the actively transcribed genes have a HNF-4α response element. These regulated genes are largely involved in the hepatic gluconeogenic program and lipid metabolism. In the pancreas HNF-4α is also a master regulator, controlling an estimated 11% of islet genes. HNF-4α protein mutations are linked to maturity-onset diabetes of the young, type 1 (MODY1) and hyperinsulinaemic hypoglycaemia. Previous structural analyses of NRs, although productive in elucidating the structure of individual domains, have lagged behind in revealing the connectivity patterns of NR domains. Here we describe the 2.9 Å crystal structure of the multidomain human HNF-4α homodimer bound to its DNA response element and coactivator-derived peptides. A convergence zone connects multiple receptor domains in an asymmetric fashion, joining distinct elements from each monomer. An arginine target of PRMT1 methylation protrudes directly into this convergence zone and sustains its integrity. A serine target of protein kinase C is also responsible for maintaining domain-domain interactions. These post-translational modifications lead to changes in DNA binding by communicating through the tightly connected surfaces of the quaternary fold. We find that some MODY1 mutations, positioned on the ligand-binding domain and hinge regions of the receptor, compromise DNA binding at a distance by communicating through the interjunctional surfaces of the complex. The overall domain representation of the HNF-4α homodimer is different from that of the PPAR-γ-RXR-α heterodimer, even when both NR complexes are assembled on the same DNA element. Our findings suggest that unique quaternary folds and interdomain connections in NRs could be exploited by small-molecule allosteric modulators that affect distal functions in these polypeptides.
Organizational Affiliation:
Metabolic Signaling and Disease Program, Sanford-Burnham Medical Research Institute, Orlando, Florida 32827, USA.