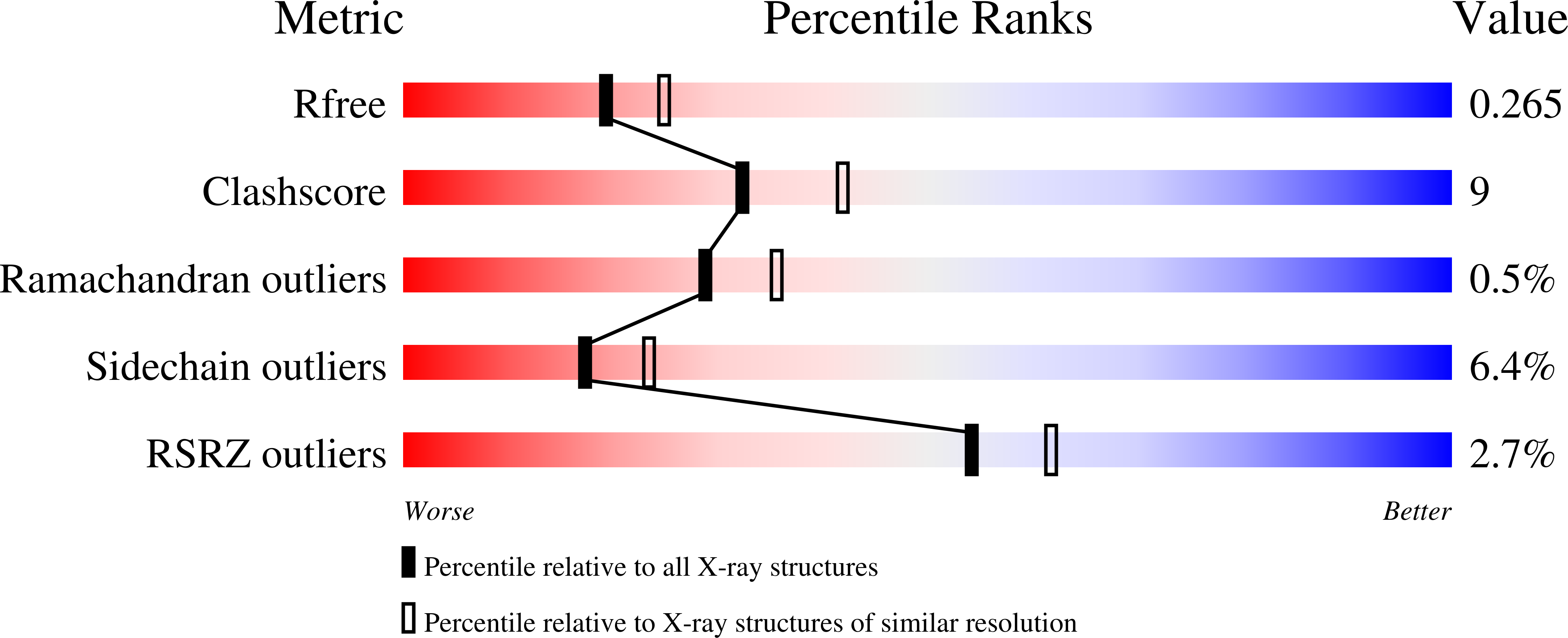
Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Shaik, M.M., Cendron, L., Salamina, M., Ruzzene, M., Zanotti, G.(2014) Mol Microbiol 91: 724-735
- PubMed: 24330328
- DOI: https://doi.org/10.1111/mmi.12487
- Primary Citation of Related Structures:
4INO, 4INP, 4LS3 - PubMed Abstract:
In Gram-negative bacteria, nickel uptake is guaranteed by multiple and complex systems that operate at the membrane and periplasmic level. Helicobacter pylori employs other yet uncharacterized systems to import the nickel required for the maturation of key enzymes, such as urease and hydrogenase. H. pylori CeuE protein (HP1561), previously annotated as the periplasmic component of an ATP-binding cassette (ABC)-type transporter apparatus responsible of haem/siderophores or other Fe(III)-complexes uptake, has been recently proposed to be on the contrary involved in nickel/cobalt acquisition. In this work, the crystal structure of H. pylori CeuE has been determined at 1.65 Å resolution using the single anomalous dispersion (SAD) method. It comprises two structurally similar globular domains, each consisting of a central five-stranded β-sheet surrounded by α-helices, an arrangement commonly classified as a Rossmann-like fold. Structurally, H. pylori CeuE belongs to the class III periplasmic substrate-binding protein. Both crystallographic data and fluorescence binding assays allow to exclude a role of the protein in the transport of Vitamin B12, enterobactin, haem and isolated Ni(2+) ions. On the contrary, the crystal structure and plasmon resonance studies about CeuE/Ni-(l-His)2 complex indicate that in H. pylori nickel transport is supported by CeuE protein and requires the presence of a natural nickelophore, analogously to what has been recently demonstrated for NikA from Escherichia coli.
Organizational Affiliation:
Department of Biomedical Sciences, University of Padua, Viale G. Colombo 3, 35131, Padua, Italy.