Structural basis for the recognition of tyrosine-based sorting signals by the mu 3A subunit of the AP-3 adaptor complex.
Mardones, G.A., Burgos, P.V., Lin, Y., Kloer, D.P., Magadan, J.G., Hurley, J.H., Bonifacino, J.S.(2013) J Biol Chem 288: 9563-9571
- PubMed: 23404500
- DOI: https://doi.org/10.1074/jbc.M113.450775
- Primary Citation of Related Structures:
4IKN - PubMed Abstract:
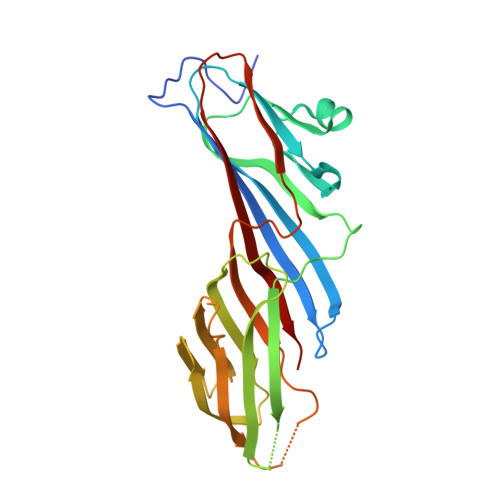
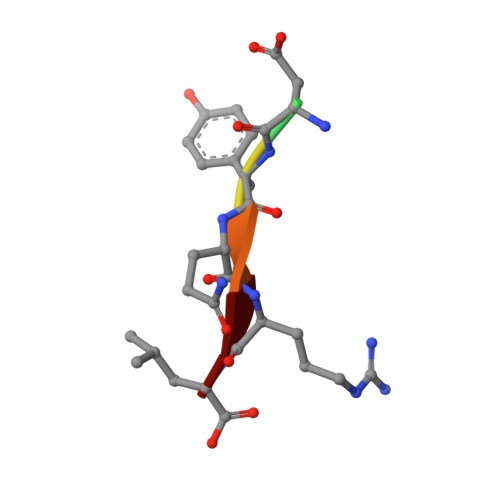
Tyrosine-based signals fitting the YXXØ motif mediate sorting of transmembrane proteins to endosomes, lysosomes, the basolateral plasma membrane of polarized epithelial cells, and the somatodendritic domain of neurons through interactions with the homologous μ1, μ2, μ3, and μ4 subunits of the corresponding AP-1, AP-2, AP-3, and AP-4 complexes. Previous x-ray crystallographic analyses identified distinct binding sites for YXXØ signals on μ2 and μ4, which were located on opposite faces of the proteins. To elucidate the mode of recognition of YXXØ signals by other members of the μ family, we solved the crystal structure at 1.85 Å resolution of the C-terminal domain of the μ3 subunit of AP-3 (isoform A) in complex with a peptide encoding a YXXØ signal (SDYQRL) from the trans-Golgi network protein TGN38. The μ3A C-terminal domain consists of an immunoglobulin-like β-sandwich organized into two subdomains, A and B. The YXXØ signal binds in an extended conformation to a site on μ3A subdomain A, at a location similar to the YXXØ-binding site on μ2 but not μ4. The binding sites on μ3A and μ2 exhibit similarities and differences that account for the ability of both proteins to bind distinct sets of YXXØ signals. Biochemical analyses confirm the identification of the μ3A site and show that this protein binds YXXØ signals with 14-19 μm affinity. The surface electrostatic potential of μ3A is less basic than that of μ2, in part explaining the association of AP-3 with intracellular membranes having less acidic phosphoinositides.
Organizational Affiliation:
Instituto de Fisiología, Facultad de Medicina, and Centro de Investigación Sur-Austral en Enfermedades del Sistema Nervioso, Universidad Austral de Chile, Valdivia 5110566, Chile.