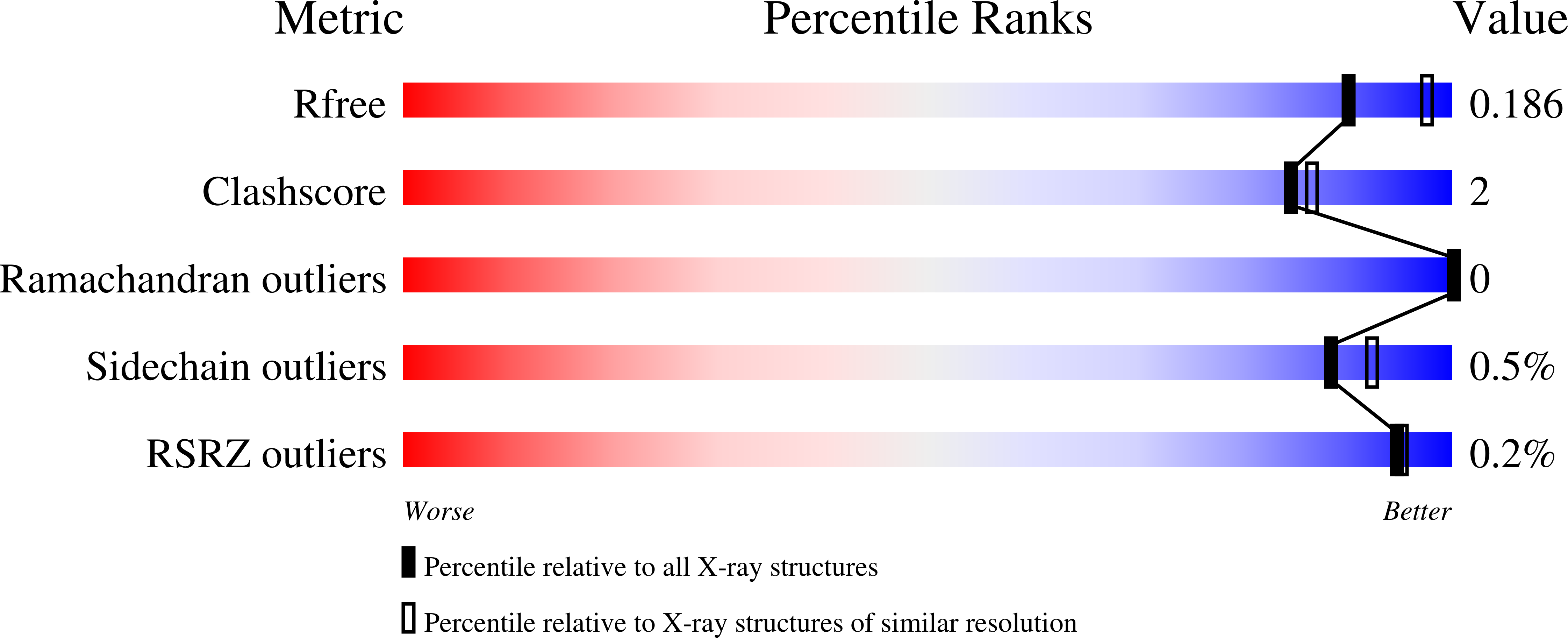
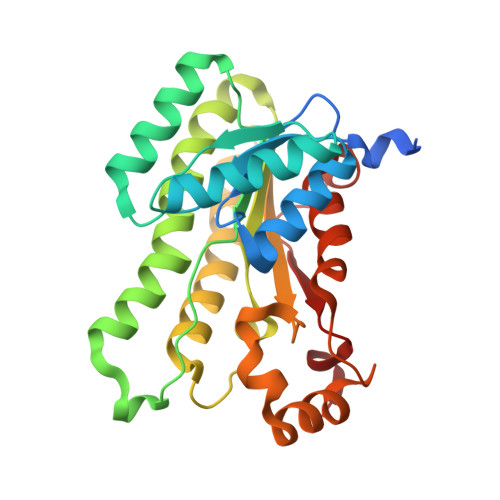
Crystal structure of a putative gluconate dehydrogenase from agrobacterium tumefaciens (target EFI-506446)
Vetting, M.W., Bouvier, J.T., Groninger-Poe, F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Imker, H.J., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.