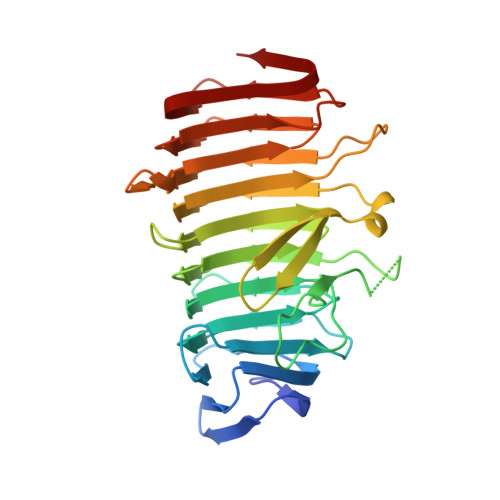
Structure of the secretion domain of HxuA from Haemophilus influenzae.
Baelen, S., Dewitte, F., Clantin, B., Villeret, V.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 1322-1327
- PubMed: 24316822
- DOI: https://doi.org/10.1107/S174430911302962X
- Primary Citation of Related Structures:
4I84 - PubMed Abstract:
Haemophilus influenzae HxuA is a cell-surface protein with haem-haemopexin binding activity which is key to haem acquisition from haemopexin and thus is one of the potential sources of haem for this microorganism. HxuA is secreted by its specific transporter HxuB. HxuA/HxuB belongs to the so-called two-partner secretion systems (TPSs) that are characterized by a conserved N-terminal domain in the secreted protein which is essential for secretion. Here, the 1.5 Å resolution structure of the secretion domain of HxuA, HxuA301, is reported. The structure reveals that HxuA301 folds into a β-helix domain with two extra-helical motifs, a four-stranded β-sheet and an N-terminal cap. Comparisons with other structures of TpsA secretion domains are reported. They reveal that despite limited sequence identity, strong structural similarities are found between the β-helix motifs, consistent with the idea that the TPS domain plays a role not only in the interaction with the specific TpsB partners but also as the scaffold initiating progressive folding of the TpsA proteins at the bacterial surface.
Organizational Affiliation:
Institut de Recherche Interdisciplinaire, IRI USR 3078 CNRS-Université Lille Nord de France, Parc CNRS de la Haute Borne, 50 Avenue de Halley, 59658 Villeneuve d'Ascq, France.