Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures.
Wu, I., Arnold, F.H.(2013) Biotechnol Bioeng 110: 1874-1883
- PubMed: 23404363
- DOI: https://doi.org/10.1002/bit.24864
- Primary Citation of Related Structures:
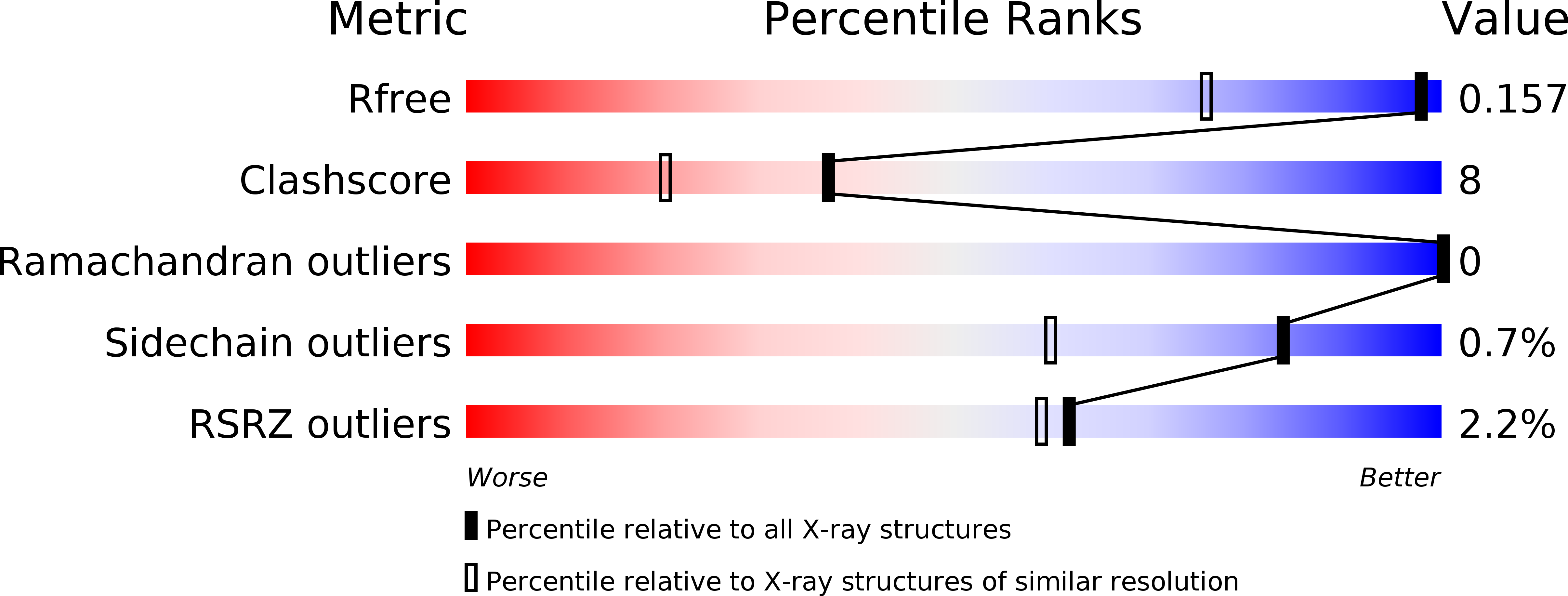
4I5R, 4I5U - PubMed Abstract:
Thermostability is an important feature in industrial enzymes: it increases biocatalyst lifetime and enables reactions at higher temperatures, where faster rates and other advantages ultimately reduce the cost of biocatalysis. Here we report the thermostabilization of a chimeric fungal family 6 cellobiohydrolase (HJPlus) by directed evolution using random mutagenesis and recombination of beneficial mutations. Thermostable variant 3C6P has a half-life of 280 min at 75°C and a T(50) of 80.1°C, a ~15°C increase over the thermostable Cel6A from Humicola insolens (HiCel6A) and a ~20°C increase over that from Hypocrea jecorina (HjCel6A). Most of the mutations also stabilize the less-stable HjCel6A, the wild-type Cel6A closest in sequence to 3C6P. During a 60-h Avicel hydrolysis, 3C6P released 2.4 times more cellobiose equivalents at its optimum temperature (T(opt)) of 75°C than HiCel6A at its T(opt) of 60°C. The total cellobiose equivalents released by HiCel6A at 60°C after 60 h is equivalent to the total released by 3C6P at 75°C after ~6 h, a 10-fold reduction in hydrolysis time. A binary mixture of thermostable Cel6A and Cel7A hydrolyzes Avicel synergistically and released 1.8 times more cellobiose equivalents than the wild-type mixture, both mixtures assessed at their respective T(opt). Crystal structures of HJPlus and 3C6P, determined at 1.5 and 1.2 Å resolution, indicate that the stabilization comes from improved hydrophobic interactions and restricted loop conformations by introduced proline residues.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology 210-41, Pasadena, CA 91125, USA.