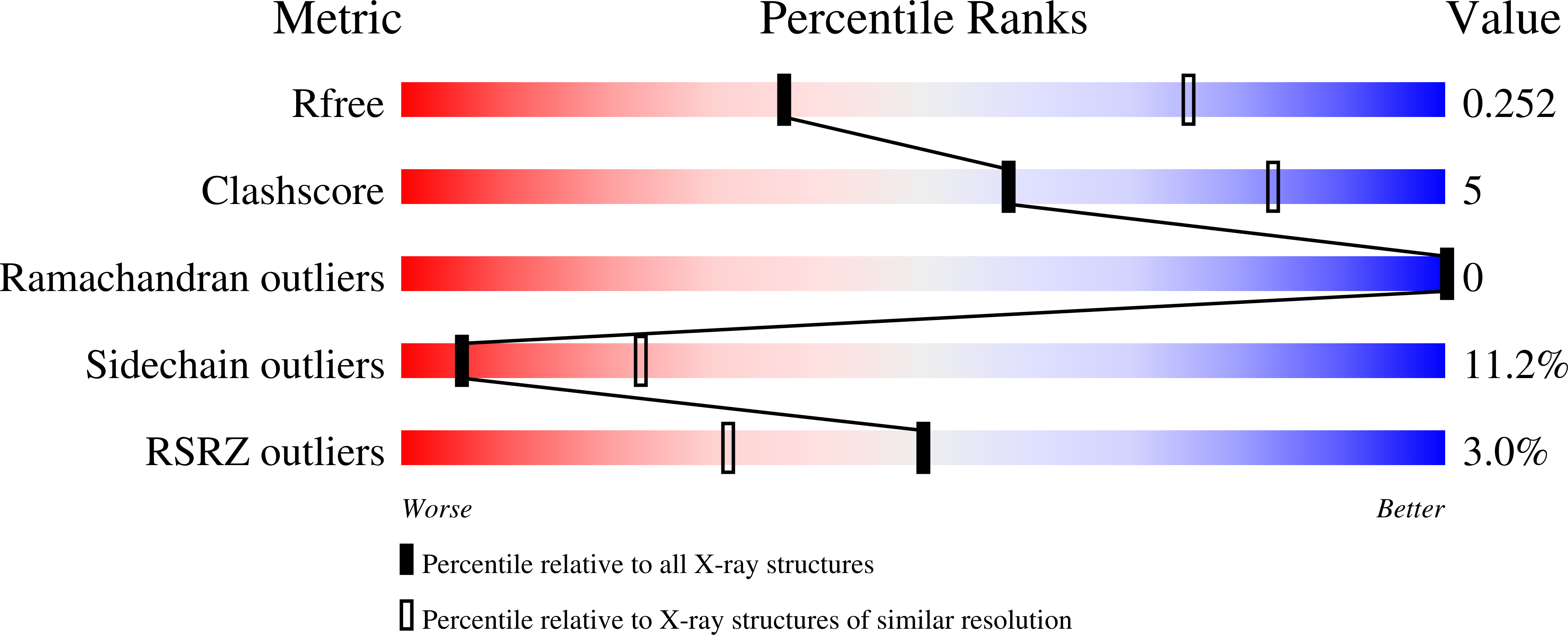
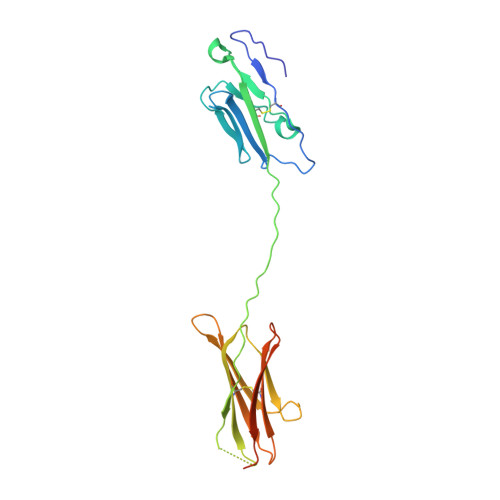
Structure and T cell inhibition properties of B7 family member, B7-H3.
Vigdorovich, V., Ramagopal, U.A., Lazar-Molnar, E., Sylvestre, E., Lee, J.S., Hofmeyer, K.A., Zang, X., Nathenson, S.G., Almo, S.C.(2013) Structure 21: 707-717
- PubMed: 23583036
- DOI: https://doi.org/10.1016/j.str.2013.03.003
- Primary Citation of Related Structures:
4I0K - PubMed Abstract:
T cell activity is controlled by a combination of antigen-dependent signaling through the T cell receptor and a set of auxiliary signals delivered through antigen-independent interactions, including the recognition of the B7 family of ligands. B7-H3 is a recently identified B7 family member that is strongly overexpressed in a range of cancers and correlates with poor prognosis. We report the crystal structure of murine B7-H3 at a 3 Å resolution, which provides a model for the organization of the IgV and IgC domains within the ectodomain. We demonstrate that B7-H3 inhibits T cell proliferation and show that the FG loop of the IgV domain plays a critical role in this function. B7-H3 crystallized as an unusual dimer arising from the exchange of the G strands in the IgV domains of partner molecules. This arrangement, in combination with previous reports, highlights the dynamic nature and plasticity of the immunoglobulin fold.
Organizational Affiliation:
Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY 10461, USA. vladimir.vigdorovich@gmail.com