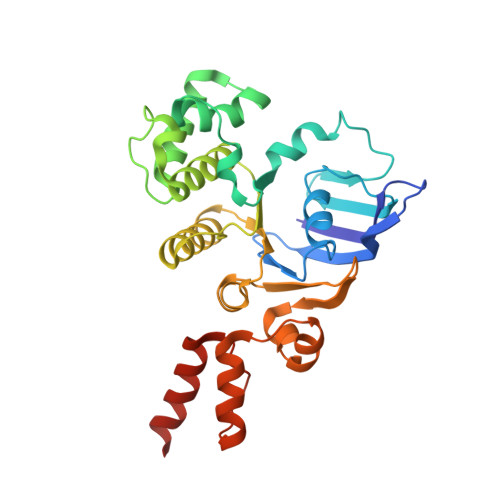
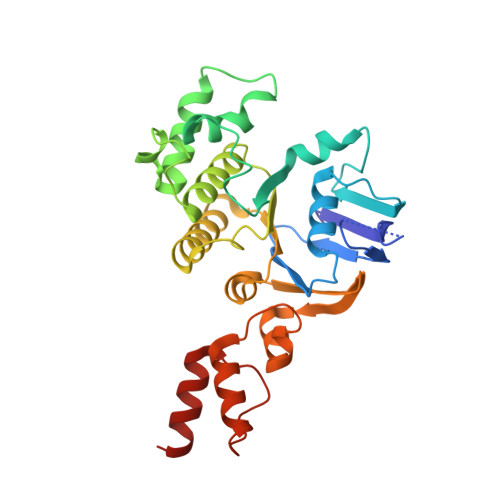
Structure of a bacterial energy-coupling factor transporter.
Wang, T., Fu, G., Pan, X., Wu, J., Gong, X., Wang, J., Shi, Y.(2013) Nature 497: 272-276
- PubMed: 23584587
- DOI: https://doi.org/10.1038/nature12045
- Primary Citation of Related Structures:
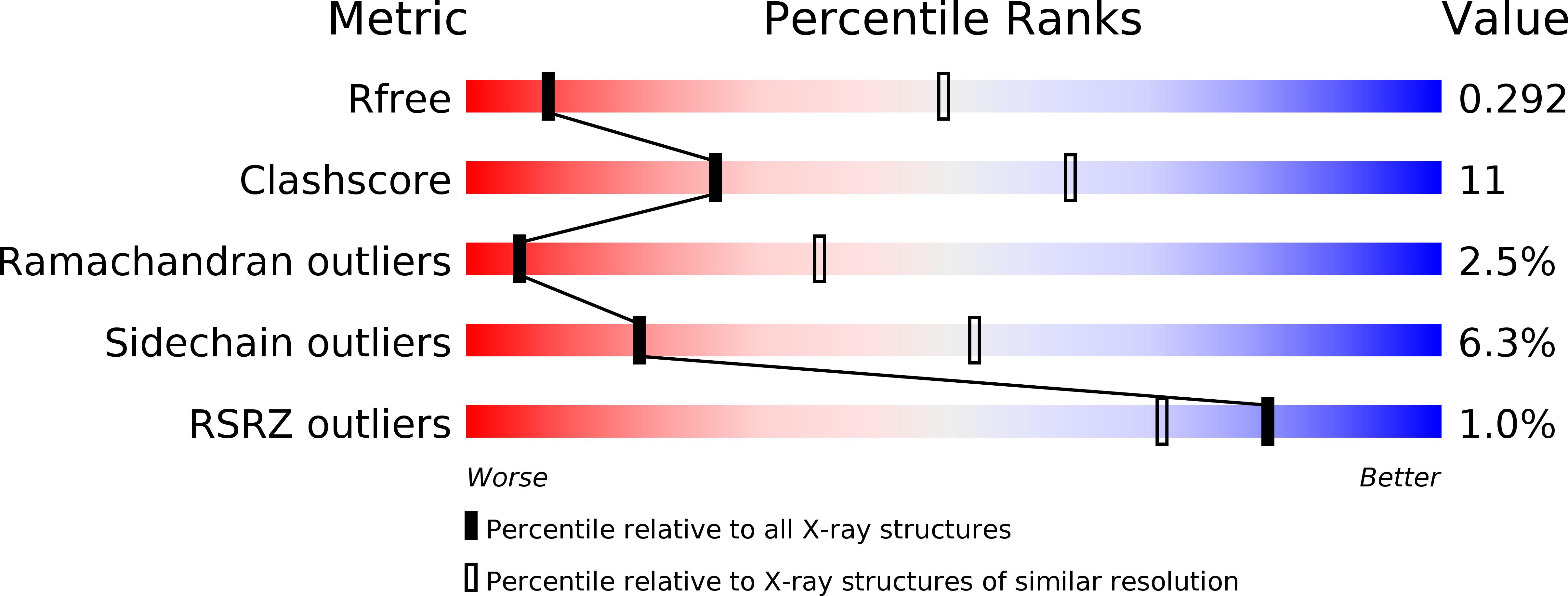
4HZU - PubMed Abstract:
The energy-coupling factor (ECF) transporters constitute a novel family of conserved membrane transporters in prokaryotes that have a similar domain organization to the ATP-binding cassette transporters. Each ECF transporter comprises a pair of cytosolic ATPases (the A and A' components, or EcfA and EcfA'), a membrane-embedded substrate-binding protein (the S component, or EcfS) and a transmembrane energy-coupling component (the T component, or EcfT) that links the EcfA-EcfA' subcomplex to EcfS. The structure and transport mechanism of the quaternary ECF transporter remain largely unknown. Here we report the crystal structure of a nucleotide-free ECF transporter from Lactobacillus brevis at a resolution of 3.5 Å. The T component has a horseshoe-shaped open architecture, with five α-helices as transmembrane segments and two cytoplasmic α-helices as coupling modules connecting to the A and A' components. Strikingly, the S component, thought to be specific for hydroxymethyl pyrimidine, lies horizontally along the lipid membrane and is bound exclusively by the five transmembrane segments and the two cytoplasmic helices of the T component. These structural features suggest a plausible working model for the transport cycle of the ECF transporters.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Science, Tsinghua University, Beijing 100084, China.