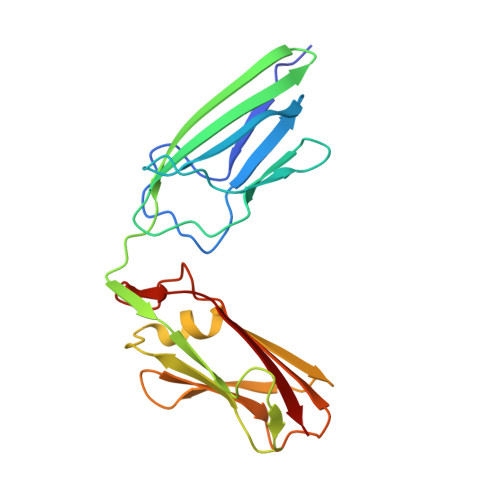
Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Both, D., Steiner, E.M., Stadler, D., Lindqvist, Y., Schnell, R., Schneider, G.(2013) Acta Crystallogr D Biol Crystallogr 69: 432-441
- PubMed: 23519418
- DOI: https://doi.org/10.1107/S0907444912049268
- Primary Citation of Related Structures:
4HU2, 4HUC - PubMed Abstract:
The transpeptidase LtdMt2 catalyzes the formation of the (3-3) cross-links characteristic of the peptidoglycan layer in the Mycobacterium tuberculosis cell wall. Bioinformatics analysis suggests that the extramembrane part of the enzyme consists of three domains: two smaller domains (denoted as A and B domains) and a transpeptidase domain (the C domain) at the C-terminus. The crystal structures of two fragments comprising the AB domains and the BC domains have been determined. The structure of the BC module, which was determined to 1.86 Å resolution using Se-SAD phasing, consists of the B domain with an immunoglobulin-related fold and the catalytic domain belonging to the ErfK/YbiS/YbnG fold family. The structure of the AB-domain fragment, which was solved by molecular replacement to 1.45 Å resolution, reveals that despite a lack of overall sequence identity the A domain is structurally very similar to the B domain. Combining the structures of the two fragments provides a view of the complete three-domain extramembrane part of LdtMt2 and shows that the protein extends at least 80-100 Å from the plasma membrane into the peptidoglycan layer and thus defines the maximal distance at which cross-links are formed by this enzyme. The LdtMt-related transpeptidases contain one or two immunoglobulin domains, which suggests that these might serve as extender units to position the catalytic domain at an appropriate distance from the membrane in the peptidoglycan layer.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institutet, S-17 177 Stockholm, Sweden.