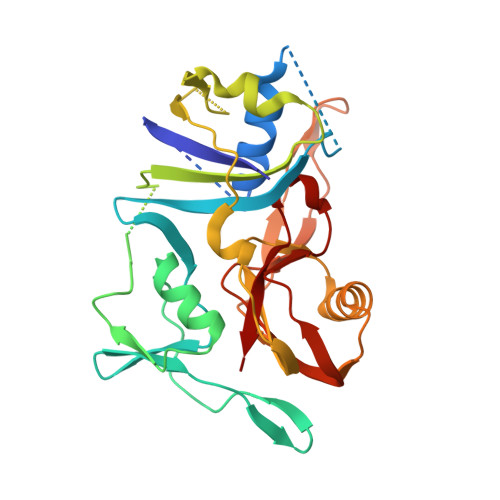
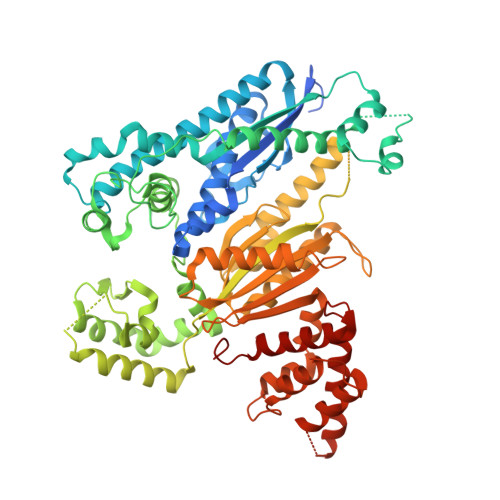
Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex.
Shao, Y., Cocozaki, A.I., Ramia, N.F., Terns, R.M., Terns, M.P., Li, H.(2013) Structure 21: 376-384
- PubMed: 23395183
- DOI: https://doi.org/10.1016/j.str.2013.01.002
- Primary Citation of Related Structures:
4H4K - PubMed Abstract:
The Cmr complex is an RNA-guided effector complex that cleaves invader RNA in the prokaryotic immune response mediated by the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat)-Cas system. Here, we report the crystal structure of a Cmr subcomplex containing Cmr2 (Cas10) and Cmr3 subunits at 2.8 Å resolution. The structure revealed a dual ferredoxin fold and glycine-rich loops characteristic of previously known repeat-associated mysterious proteins and two unique insertion elements in Cmr3 that mediate its interaction with Cmr2. Surprisingly, while mutation of both insertion elements significantly weakened Cmr3-Cmr2 interaction, they exhibit differential effects on Cmr-mediated RNA cleavage by the Cmr complex, suggesting stabilization of Cmr2-Cmr3 interactions by other subunits. Further mutational analysis of the two conserved (but non-Cmr2-binding) glycine-rich loops of Cmr3 identified a region that is likely involved in assembly or the RNA cleavage function of the Cmr complex.
Organizational Affiliation:
Institute of Molecular Biophysics, Florida State University, Tallahassee, FL 32306, USA.