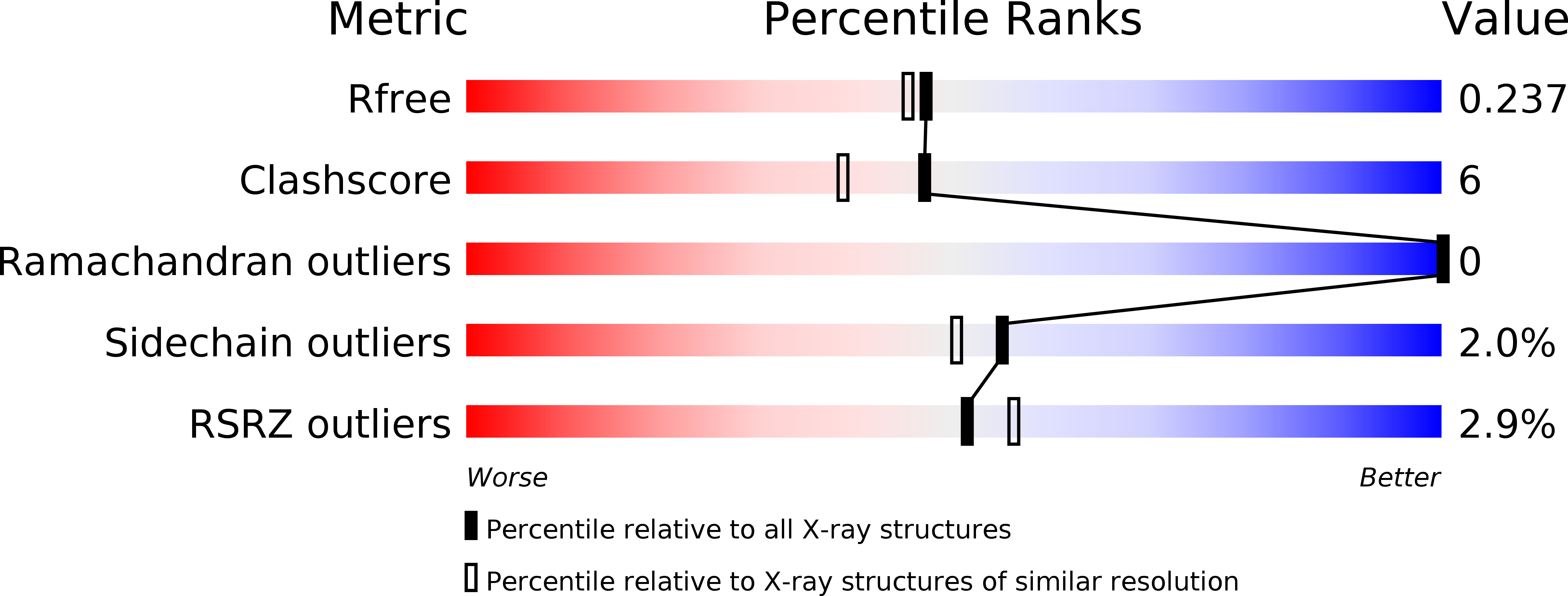
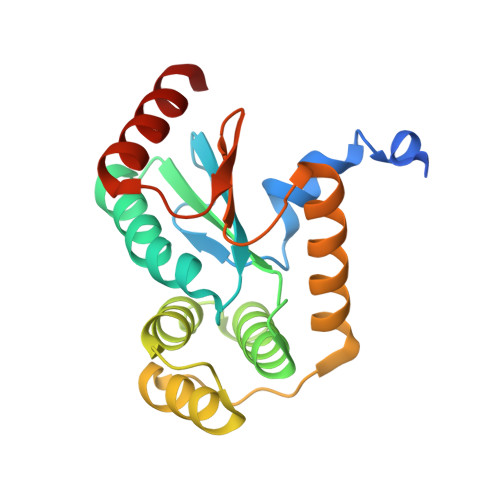
Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium
Shepherd, M., Heras, B., Achard, M.E., King, G.J., Argente, M.P., Kurth, F., Taylor, S.L., Howard, M.J., King, N.P., Schembri, M.A., McEwan, A.G.(2013) Antioxid Redox Signal 19: 1494-1506
- PubMed: 23642141
- DOI: https://doi.org/10.1089/ars.2012.4939
- Primary Citation of Related Structures:
4GXZ - PubMed Abstract:
The prototypical protein disulfide bond (Dsb) formation and protein refolding pathways in the bacterial periplasm involving Dsb proteins have been most comprehensively defined in Escherichia coli. However, genomic analysis has revealed several distinct Dsb-like systems in bacteria, including the pathogen Salmonella enterica serovar Typhimurium. This includes the scsABCD locus, which encodes a system that has been shown via genetic analysis to confer copper tolerance, but whose biochemical properties at the protein level are not defined. The aim of this study was to provide functional insights into the soluble ScsC protein through structural, biochemical, and genetic analyses.
Organizational Affiliation:
1 School of Biosciences, University of Kent , Canterbury, United Kingdom .