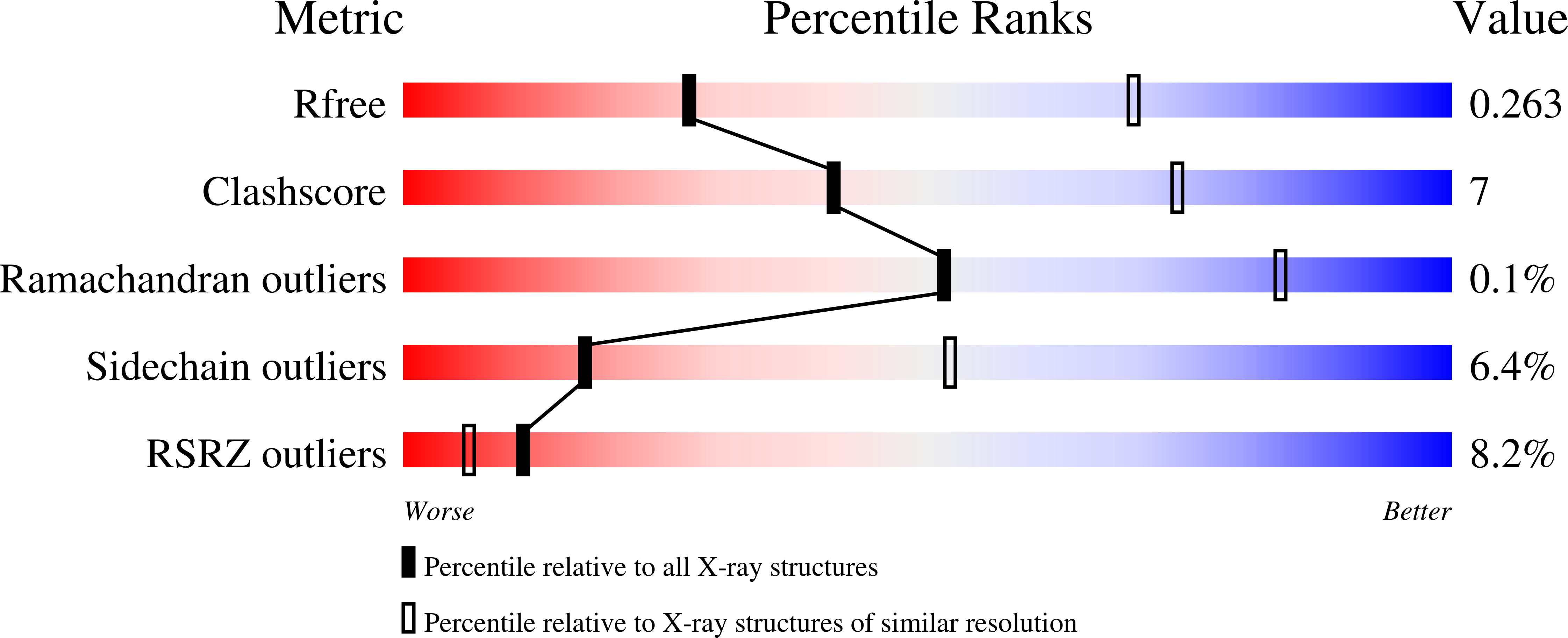
Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
Kong, C., Zeng, W., Ye, S., Chen, L., Sauer, D.B., Lam, Y., Derebe, M.G., Jiang, Y.(2012) Elife 1: e00184-e00184
- PubMed: 23240087
- DOI: https://doi.org/10.7554/eLife.00184
- Primary Citation of Related Structures:
4GVL, 4GX0, 4GX1, 4GX2, 4GX5 - PubMed Abstract:
The gating ring-forming RCK domain regulates channel gating in response to various cellular chemical stimuli in eukaryotic Slo channel families and the majority of ligand-gated prokaryotic K(+) channels and transporters. Here we present structural and functional studies of a dual RCK-containing, multi-ligand gated K(+) channel from Geobacter sulfurreducens, named GsuK. We demonstrate that ADP and NAD(+) activate the GsuK channel, whereas Ca(2+) serves as an allosteric inhibitor. Multiple crystal structures elucidate the structural basis of multi-ligand gating in GsuK, and also reveal a unique ion conduction pore with segmented inner helices. Structural comparison leads us to propose a novel pore opening mechanics that is distinct from other K(+) channels.DOI:http://dx.doi.org/10.7554/eLife.00184.001.
Organizational Affiliation:
Department of Physiology , University of Texas Southwestern Medical Center , Dallas , United States.